A single-cell, spatial transcriptomic atlas of the Arabidopsis life cycle
IF 13.6
1区 生物学
Q1 PLANT SCIENCES
引用次数: 0
Abstract
Arabidopsis has been pivotal in uncovering fundamental principles of plant biology, yet a comprehensive, high-resolution understanding of its cellular identities throughout the entire life cycle remains incomplete. Here we present a single-nucleus and spatial transcriptomic atlas spanning ten developmental stages, encompassing over 400,000 nuclei from all organ systems and tissues—from seeds to developing siliques. Leveraging paired single-nucleus and spatial transcriptomic datasets, we annotate 75% of identified cell clusters, revealing striking molecular diversity in cell types and states across development. Our integrated approach identified conserved transcriptional signatures among recurrent cell types, organ-specific heterogeneity and previously uncharacterized cell-type-specific markers validated spatially. Moreover, we uncover dynamic transcriptional programs governing secondary metabolite production and differential growth patterns, exemplified by detailed spatial profiling of the compact yet complex apical hook structure; this profiling revealed transient cellular states linked to developmental progression and hormonal regulation, highlighting the hidden complexity underlying plant morphogenesis. Functional validation of genes uniquely expressed within specific cell contexts confirmed their essential developmental roles, underscoring the predictive power of our atlas. Collectively, this comprehensive resource provides an invaluable foundation for exploring cellular differentiation, environmental responses and genetic perturbations at high resolution, advancing our understanding of plant biology. This study presents an extensive single-nucleus and spatial transcriptomic atlas of the Arabidopsis life cycle that represents ten distinct developmental time points inclusive of six diverse organs.
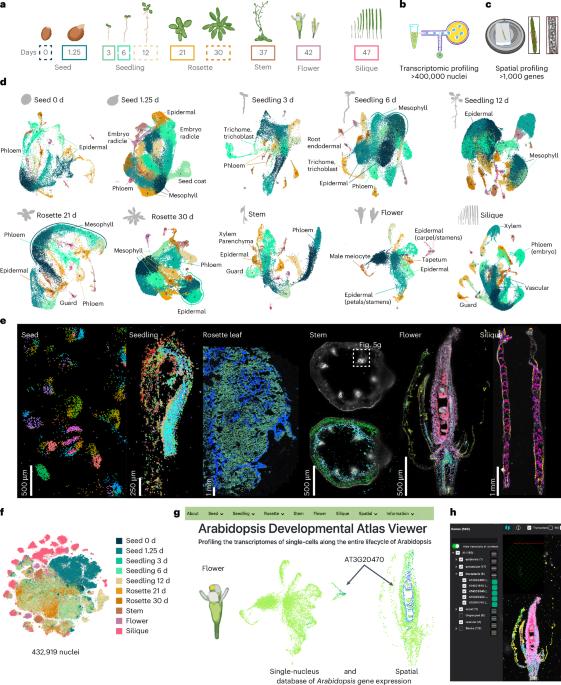
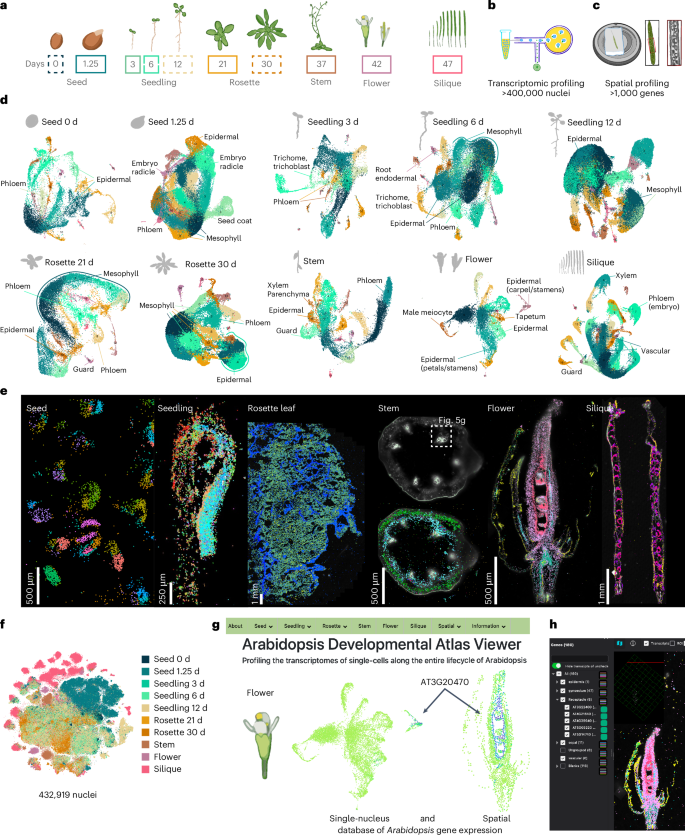
拟南芥生命周期的单细胞空间转录组图谱。
拟南芥在揭示植物生物学的基本原理方面起着关键作用,但对其整个生命周期的细胞身份的全面、高分辨率理解仍不完整。在这里,我们展示了一个跨越10个发育阶段的单核和空间转录组图谱,包括来自所有器官系统和组织的400,000多个细胞核——从种子到发育中的硅片。利用配对的单核和空间转录组数据集,我们注释了75%的已鉴定细胞簇,揭示了细胞类型和发育状态的惊人分子多样性。我们的综合方法确定了复发细胞类型、器官特异性异质性和先前未表征的细胞类型特异性标记物之间的保守转录特征。此外,我们发现了控制次生代谢物产生和差异生长模式的动态转录程序,例如紧凑而复杂的顶钩结构的详细空间剖面;这一分析揭示了与发育过程和激素调节相关的短暂细胞状态,突出了植物形态发生背后隐藏的复杂性。在特定细胞环境中唯一表达的基因的功能验证证实了它们的重要发育作用,强调了我们的图谱的预测能力。总的来说,这一综合资源为高分辨率探索细胞分化、环境反应和遗传扰动提供了宝贵的基础,促进了我们对植物生物学的理解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Plants
PLANT SCIENCES-
CiteScore
25.30
自引率
2.20%
发文量
196
期刊介绍:
Nature Plants is an online-only, monthly journal publishing the best research on plants — from their evolution, development, metabolism and environmental interactions to their societal significance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


