AlphaCD: a machine learning model capable of highly accurate characterization for 21,335 cytidine deaminases
IF 25.9
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
The vast scope but limited-supporting evidence in sequence databases hinders identification of proteins with specific functionality. Here, we experimentally characterized catalytic efficiency, target site window, motif preference, and off-target activity of 1100 apolipoprotein B mRNA-editing enzyme, catalytic polypeptide (APOBEC)-like family cytidine deaminases (CDs) fused with nCas9 in HEK293T cells, thereby generating the largest dataset of experimentally validated functions for a single protein family to date. These data, together with amino acid sequence, three-dimensional structure, and eight additional features, were used to construct a machine learning (ML) model, AlphaCD, which showed high accuracy in predicting catalytic efficiency (0.92) and off-target activity (0.84), as well as target windows (0.73) and catalytic motifs (0.78). We applied the trained model to predict the above catalytic features of 21,335 CDs in Uniprot, and subsampling of 28 CDs further validated its prediction accuracy (0.84, 0.87, 0.75, 0.73, respectively). Alanine scanning-based mutagenesis was then employed to reduce off-targets in one example CD, which produced a remarkably high fidelity, high efficiency cytosine base editor, thus demonstrating AlphaCD application in high-accuracy, high-throughput protein functional characterization, and providing a strategy for accelerated characterization of other proteins.
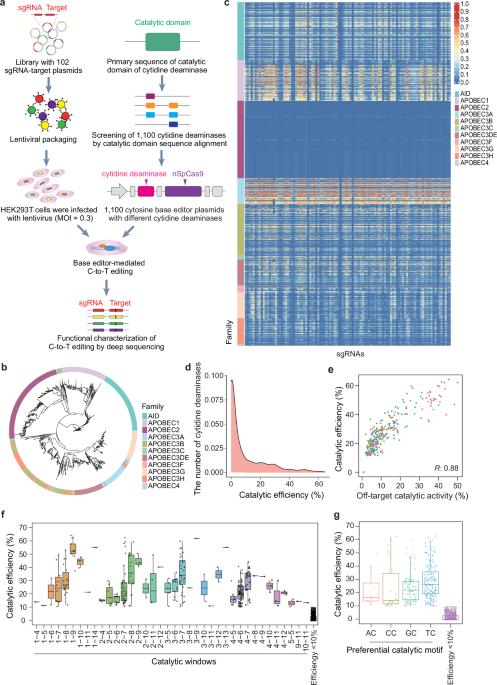
alphaacd:一种机器学习模型,能够高度准确地表征21,335种胞苷脱氨酶。
序列数据库中范围广泛但支持证据有限,阻碍了具有特定功能的蛋白质的鉴定。在这里,我们通过实验表征了1100种载脂蛋白B mrna编辑酶、催化多肽(APOBEC)样家族胞苷脱氨酶(CDs)在HEK293T细胞中与nCas9融合的催化效率、靶位窗口、基元偏好和脱靶活性,从而生成了迄今为止单个蛋白质家族实验验证功能的最大数据集。这些数据与氨基酸序列、三维结构和8个附加特征一起用于构建机器学习(ML)模型AlphaCD,该模型在预测催化效率(0.92)和脱靶活性(0.84)以及目标窗口(0.73)和催化基序(0.78)方面具有很高的准确性。我们应用训练好的模型预测了Uniprot中21335个cd的上述催化特征,并对28个cd进行了抽样,进一步验证了其预测精度(分别为0.84、0.87、0.75、0.73)。然后利用基于丙氨酸扫描的诱变技术减少了一个例子CD中的脱靶,产生了一个非常高保真度、高效率的胞嘧啶碱基编辑器,从而证明了AlphaCD在高精度、高通量蛋白质功能表征中的应用,并为其他蛋白质的加速表征提供了一种策略。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cell Research
生物-细胞生物学
CiteScore
53.90
自引率
0.70%
发文量
2420
审稿时长
2.3 months
期刊介绍:
Cell Research (CR) is an international journal published by Springer Nature in partnership with the Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences (CAS). It focuses on publishing original research articles and reviews in various areas of life sciences, particularly those related to molecular and cell biology. The journal covers a broad range of topics including cell growth, differentiation, and apoptosis; signal transduction; stem cell biology and development; chromatin, epigenetics, and transcription; RNA biology; structural and molecular biology; cancer biology and metabolism; immunity and molecular pathogenesis; molecular and cellular neuroscience; plant molecular and cell biology; and omics, system biology, and synthetic biology. CR is recognized as China's best international journal in life sciences and is part of Springer Nature's prestigious family of Molecular Cell Biology journals.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


