Transcriptomic Analysis Reveals the Key Regulatory Pathways of the Gills and Liver of Grass Carp Under High NaHCO3Stress
Abstract
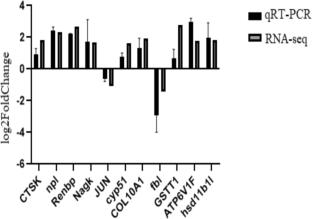
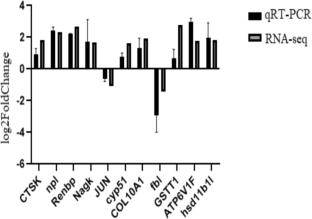
To elucidate the molecular mechanisms by which grass carp respond to high-concentration NaHCO3 stress, this study employed RNA-sequencing technology to perform transcriptome analysis on the gill and liver tissues of the treatment group with 30 mmol/L NaHCO3 and the control group (0 mmol/L). Through sequencing of 12 libraries constructed from 6 gill samples and 6 liver samples, a total of 41.86 GB of high-quality data from gill tissues and 41.49 GB of high-quality data from liver tissues were obtained. The analysis revealed that there were 1223 and 439 significantly differentially expressed genes (DEGs) in the gill and liver tissues, respectively, under NaHCO3 stress. Functional enrichment (GO) analysis indicated that grass carp respond to alkaline stress by regulating biological processes such as protein hydrolysis, ATP-binding activity, and lipid metabolism. Pathway (KEGG) analysis further revealed that immune-related signaling pathways were significantly activated to resist external stress, and the elevation of energy metabolism levels may provide necessary support for the stress-resistance process. This study systematically revealed the molecular adaptation strategies of grass carp in a high-alkali environment, providing an important theoretical basis for the molecular breeding of saline-alkali-tolerant grass carp varieties and the research on stress-resistance mechanisms.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


