Identification of salivary inflammatory non coding RNA biomarkers for coronary artery disease through epicardial adipose tissue gene network analysis
IF 5.7
2区 医学
Q1 CARDIAC & CARDIOVASCULAR SYSTEMS
引用次数: 0
Abstract
Background and aim
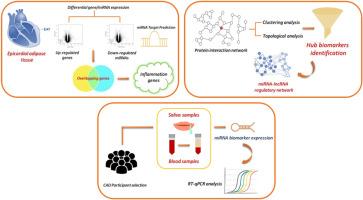
Epicardial adipose tissue (EAT), due to its proximity to the myocardium, contributes inflammatory profile in CAD onset and progression. Emerging evidence highlights the role of EAT-derived non-coding RNAs in modulating disease pathways. However, identifying EAT-specific biomarkers remains challenging for routine diagnosis. Therefore, this study aimed to identify non-invasive, multi-source biomarkers from EAT, plasma, and saliva, and their regulatory miRNAs and lncRNAs, through network analysis, regulatory mapping, and experimental validation.
Methods
Differential expression analyses identified upregulated genes and miRNAs in EAT. Protein interaction network was constructed for overlapping upregulated genes and downregulated miRNA targets along with inflammatory-related genes. Clustering, ontology, and topological analyses were performed to pinpoint hub biomarkers and regulatory non-coding RNAs. Key candidate molecules were then experimentally validated using saliva and plasma samples from a cohort of CAD patients (n = 30) and healthy controls (n = 30).
Results
A total of 840 upregulated and 1817 downregulated genes were identified in EAT with 36 upregulated and 82 downregulated miRNAs. Among 343 overexpressed inflammatory genes, CD8A, IL7R, and CCL5 emerged as hub biomarkers. Regulatory analysis uncovered 18 miRNAs and 548 lncRNAs influencing these hubs. Notably, hsa-miR-582–3p exhibited the highest number of lncRNA interactions and was significantly downregulated in saliva and plasma of CAD patients (p < 0.05).
Conclusions
Our study establishes a link between EAT inflammation and salivary expression profiles, highlighting hsa-miR-582–3p as a promising non-invasive salivary biomarker for CAD. This integrative approach provides a valuable foundation for developing multi-source diagnostics in cardiovascular clinical settings.
通过心外膜脂肪组织基因网络分析鉴定冠状动脉疾病唾液炎性非编码RNA生物标志物
背景和目的心外膜脂肪组织(EAT),由于其靠近心肌,在冠心病的发生和发展中具有炎症特征。新出现的证据强调了eat衍生的非编码rna在调节疾病通路中的作用。然而,确定eat特异性生物标志物对于常规诊断仍然具有挑战性。因此,本研究旨在通过网络分析、调控作图和实验验证,鉴定来自EAT、血浆和唾液的无创多源生物标志物及其调控mirna和lncrna。方法通过差异表达分析鉴定EAT中上调的基因和mirna。构建蛋白相互作用网络,将上调基因与下调miRNA靶点与炎症相关基因重叠。进行聚类、本体和拓扑分析,以确定中心生物标志物和调节性非编码rna。然后使用来自CAD患者(n = 30)和健康对照(n = 30)的唾液和血浆样本对关键候选分子进行实验验证。结果在EAT中共鉴定出840个上调基因和1817个下调基因,其中上调基因36个,下调基因82个。在343个过表达的炎症基因中,CD8A、IL7R和CCL5成为中枢生物标志物。调控分析发现18个mirna和548个lncrna影响这些枢纽。值得注意的是,hsa-miR-582-3p在CAD患者的唾液和血浆中表现出最多的lncRNA相互作用,并显著下调(p < 0.05)。sour研究建立了EAT炎症与唾液表达谱之间的联系,强调hsa-miR-582-3p是一种有前途的非侵入性CAD唾液生物标志物。这种综合方法为在心血管临床环境中发展多源诊断提供了宝贵的基础。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Atherosclerosis
医学-外周血管病
CiteScore
9.80
自引率
3.80%
发文量
1269
审稿时长
36 days
期刊介绍:
Atherosclerosis has an open access mirror journal Atherosclerosis: X, sharing the same aims and scope, editorial team, submission system and rigorous peer review.
Atherosclerosis brings together, from all sources, papers concerned with investigation on atherosclerosis, its risk factors and clinical manifestations. Atherosclerosis covers basic and translational, clinical and population research approaches to arterial and vascular biology and disease, as well as their risk factors including: disturbances of lipid and lipoprotein metabolism, diabetes and hypertension, thrombosis, and inflammation. The Editors are interested in original or review papers dealing with the pathogenesis, environmental, genetic and epigenetic basis, diagnosis or treatment of atherosclerosis and related diseases as well as their risk factors.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


