Profiles of HBV DNA integration in humans with hepatitis B virus infection: Insights for antiviral treatment
IF 7.5
1区 医学
Q1 GASTROENTEROLOGY & HEPATOLOGY
引用次数: 0
Abstract
Background & Aims
HBV integration profiles in the natural history of chronic HBV infection (CHB) have not been well-defined. Hence, we aimed to determine HBV integration profiles across different CHB phases.
Methods
We delineated integration profiles from liver biopsies of 55 patients in different CHB phases (3 HBsAg-positive/HBeAg-positive infection; 13 HBsAg-positive/HBeAg-positive hepatitis; 7 HBsAg-positive/HBeAg-negative infection; 12 HBsAg-positive/HBeAg-negative hepatitis; 10 HBsAg seroclearance; 10 occult HBV). Target-capture next-generation sequencing (NovaSeq-6000) was performed, and integrations were characterized on AVID (integrations defined as chimeric fusions in ≥1 soft-clipped reads and ≥2 total reads).
Results
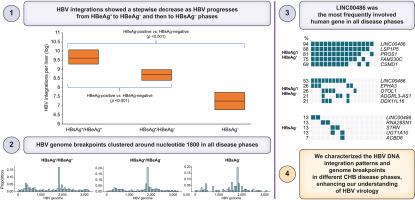
HBV integrations were detected in 35 HBsAg-positive (100%), 8 (80%) HBsAg seroclearance, and 7 (70%) occult HBV patients, respectively. There was a stepwise decrease in integration events from HBsAg-positive/HBeAg-positive (median 9.6 [IQR 9.3-10.1] log integrations per liver), HBsAg-positive/HBeAg-negative (8.7 [8.4-9.0] log integrations) and HBsAg-negative patient groups (7.3 [6.8-7.7] log integrations) (p <0.001 for trend). There were no differences in integration frequencies in chronic infection (ALT < the upper limit of normal) and chronic hepatitis (ALT ≥ the upper limit of normal) for either HBeAg-positive or -negative patients (all p >0.05). No significant differences in integration frequencies were noted between HBsAg seroclearance and occult HBV groups (p >0.05). HBV genome integration breakpoints clustered around nucleotide 1800 in all disease phases. Human genome breakpoints were also delineated, and LINC00486 was the most frequently involved human gene in all disease phases.
Conclusion
We characterized the HBV DNA integration patterns and genome breakpoints of patients in different CHB disease phases. These findings enhance our understanding of the natural history of CHB.
Impact and implications
HBV integration profiles in the natural history of chronic HBV infection have not been well-defined. We utilized next-generation sequencing in a well-characterized cohort of patients with HBV at different disease phases, demonstrating a stepwise decrease in integration events as HBV progressed from HBeAg-positive to HBeAg-negative, and then to HBsAg-negative phases. Human and viral genome breakpoints were also identified. These findings enhance our understanding of the natural history of CHB, and provide insights for antiviral treatment.
乙型肝炎病毒感染者HBV DNA整合概况:抗病毒治疗的见解
背景:在慢性HBV感染(CHB)的自然史中,AimsHBV整合谱尚未明确定义。因此,我们的目的是确定HBV在不同CHB期的整合概况。方法对55例不同期CHB患者(3例HBsAg阳性/ hbeag阳性感染,13例HBsAg阳性/ hbeag阳性肝炎,7例HBsAg阳性/ hbeag阴性感染,12例HBsAg阳性/ hbeag阴性肝炎,10例HBsAg血清清除,10例隐匿性HBV)的肝脏活检进行整合分析。进行目标捕获下一代测序(NovaSeq-6000),并用AVID对整合进行表征(整合定义为在≥1个软剪切reads和≥2个总reads中嵌合融合)。结果HBsAg阳性35例(100%)、HBsAg清除率8例(80%)、隐匿性HBV 7例(70%)分别检测到shbv整合。从hbsag阳性/ hbeag阳性患者组(平均每肝9.6 [IQR 9.3-10.1]个对数积分)、hbsag阳性/ hbeag阴性患者组(8.7[8.4-9.0]个对数积分)和hbsag阴性患者组(7.3[6.8-7.7]个对数积分)的整合事件逐步减少(趋势p <;0.001)。hbeag阳性和阴性患者的慢性感染(ALT <;正常上限)和慢性肝炎(ALT≥正常上限)积分频率无差异(p >0.05)。HBsAg清除率组与隐性HBV组的整合频率无显著差异(p >0.05)。在所有疾病阶段,HBV基因组整合断点都聚集在核苷酸1800附近。人类基因组断点也被描绘出来,LINC00486是所有疾病阶段最常涉及的人类基因。结论我们对不同CHB阶段患者的HBV DNA整合模式和基因组断点进行了分析。这些发现增强了我们对慢性乙型肝炎自然历史的认识。影响和意义慢性HBV感染自然史中的shbv整合概况尚未明确定义。我们对处于不同疾病阶段的HBV患者进行了新一代测序,结果表明,随着HBV从hbeag阳性进展到hbeag阴性,再到hbsag阴性,整合事件逐步减少。人类和病毒基因组断点也被确定。这些发现增强了我们对慢性乙型肝炎自然历史的理解,并为抗病毒治疗提供了见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

JHEP Reports
GASTROENTEROLOGY & HEPATOLOGY-
CiteScore
12.40
自引率
2.40%
发文量
161
审稿时长
36 days
期刊介绍:
JHEP Reports is an open access journal that is affiliated with the European Association for the Study of the Liver (EASL). It serves as a companion journal to the highly respected Journal of Hepatology.
The primary objective of JHEP Reports is to publish original papers and reviews that contribute to the advancement of knowledge in the field of liver diseases. The journal covers a wide range of topics, including basic, translational, and clinical research. It also focuses on global issues in hepatology, with particular emphasis on areas such as clinical trials, novel diagnostics, precision medicine and therapeutics, cancer research, cellular and molecular studies, artificial intelligence, microbiome research, epidemiology, and cutting-edge technologies.
In summary, JHEP Reports is dedicated to promoting scientific discoveries and innovations in liver diseases through the publication of high-quality research papers and reviews covering various aspects of hepatology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


