Nanobodies restore stability to cancer-associated mutants of tumor suppressor protein p16INK4a
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
We describe the generation and characterization of camelid single-domain antibodies (nanobodies) raised against tumor suppressor protein p16INK4a (p16). p16 is a cell cycle regulator that inhibits cyclin-dependent kinases CDK4 and CDK6 and is inactivated in sporadic and familial cancers. The majority of p16 missense mutations cause loss of function by destabilizing the protein’s structure. We identify nanobodies that bind p16 with nanomolar affinities and restore the stability of many different cancer-associated p16 mutations located at sites throughout the protein. The crystal structure of a nanobody-p16 complex reveals that the nanobody binds to the opposite face of p16 to the CDK-binding interface permitting formation of a ternary complex. We confirm that nanobodies bind to p16 in a cellular setting and do not preclude p16 binding to CDK6 and its ability to induce cell-cycle arrest. These findings indicate that nanobodies merit testing as pharmacological chaperones for p16 reactivation in the cell.
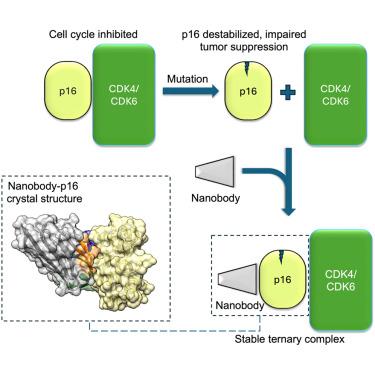
纳米体恢复肿瘤抑制蛋白p16INK4a癌症相关突变体的稳定性
我们描述了针对肿瘤抑制蛋白p16INK4a (p16)的骆驼单域抗体(纳米体)的产生和表征。p16是一种细胞周期调节因子,抑制细胞周期蛋白依赖性激酶CDK4和CDK6,在散发性和家族性癌症中失活。大多数p16错义突变通过破坏蛋白质结构的稳定而导致功能丧失。我们发现了具有纳米摩尔亲和力的结合p16的纳米体,并恢复了位于整个蛋白质位点的许多不同癌症相关p16突变的稳定性。纳米体-p16复合物的晶体结构表明,纳米体结合在p16与cdk结合界面的相反面,从而形成三元配合物。我们证实纳米体在细胞环境中与p16结合,并不妨碍p16与CDK6的结合及其诱导细胞周期阻滞的能力。这些发现表明纳米体值得作为细胞中p16再激活的药理学伴侣进行测试。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


