Mouse trophectoderm stem cells generated with morula signalling inducers capture an early trophectoderm state
IF 19.1
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
The first embryonic cell differentiation in mice segregates the trophectoderm and the inner cell mass. Successful derivation of mouse trophoblast stem cells (TSCs) and trophectoderm stem cells (TESCs) has greatly facilitated the understanding of trophoblast differentiation. However, our understanding of early trophectoderm differentiation remains incomplete. Here we report the establishment of a morula-derived trophectoderm stem cell (MTSC) line from 32-cell embryos that show enhanced and uniform trophoblast core gene expression. Importantly, distinct from TSCs or TESCs, MTSCs represent a much earlier trophectoderm state (E3.5) than that of TSCs (E5.5–6.5) and TESCs (E4.5–5.5). MTSCs can robustly integrate into all cell lineages of the placenta. Moreover, MTSCs can self-organize to form placenta organoids. When partially differentiated MTSCs aggregate with embryonic stem cells, they form blastoids that efficiently implant uteruses. Finally, MSTC medium can efficiently convert embryonic stem cells, TSCs and TESCs into MTSC-like cells. Thus, MTSCs capture an early blastocyst trophectoderm state and provide a research model for studying trophoblast development. Gao, Li and colleagues derive trophectoderm stem cells from 32-cell mouse embryos. These cells represent an early trophectoderm state and are capable of developing into placenta cells, forming placental organoids and contributing to blastoid generation.
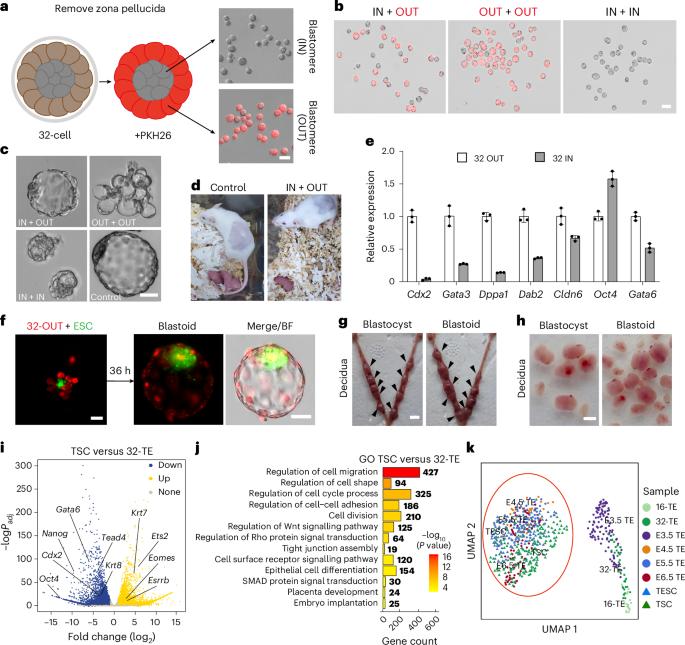
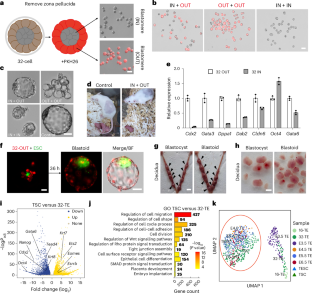
用桑葚信号诱导剂生成的小鼠滋养外胚层干细胞捕获早期滋养外胚层状态
小鼠第一次胚胎细胞分化分化为滋养外胚层和内细胞团。小鼠滋养层干细胞(TSCs)和滋养外胚层干细胞(TESCs)的成功衍生极大地促进了对滋养层分化的理解。然而,我们对早期滋养外胚层分化的理解仍然不完整。在这里,我们报告了从32个细胞胚胎中建立的桑葚来源的滋养外胚层干细胞(MTSC)系,显示出增强和均匀的滋养层核心基因表达。重要的是,与TSCs或TESCs不同,MTSCs比TSCs (E5.5-6.5)和TESCs (E4.5-5.5)表现出更早的滋养外胚层状态(E3.5)。间充质干细胞可以整合到胎盘的所有细胞系中。此外,间充质干细胞可以自组织形成胎盘类器官。当部分分化的间充质干细胞与胚胎干细胞聚集时,它们形成囊胚,有效地植入子宫。最后,MSTC培养基可以有效地将胚胎干细胞、TSCs和TESCs转化为mtsc样细胞。因此,间充质干细胞捕获早期囊胚滋养外胚层状态,为研究滋养层发育提供了一种研究模型。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Cell Biology
生物-细胞生物学
CiteScore
28.40
自引率
0.90%
发文量
219
审稿时长
3 months
期刊介绍:
Nature Cell Biology, a prestigious journal, upholds a commitment to publishing papers of the highest quality across all areas of cell biology, with a particular focus on elucidating mechanisms underlying fundamental cell biological processes. The journal's broad scope encompasses various areas of interest, including but not limited to:
-Autophagy
-Cancer biology
-Cell adhesion and migration
-Cell cycle and growth
-Cell death
-Chromatin and epigenetics
-Cytoskeletal dynamics
-Developmental biology
-DNA replication and repair
-Mechanisms of human disease
-Mechanobiology
-Membrane traffic and dynamics
-Metabolism
-Nuclear organization and dynamics
-Organelle biology
-Proteolysis and quality control
-RNA biology
-Signal transduction
-Stem cell biology
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


