Identification and isolation pipeline for single cell transcriptomic of magnetically enriched tumor cells
IF 3.7
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
Introduction
The transcriptomic analysis of circulating tumor cells provides valuable insights into cancer metastasis, heterogeneity and the identification of drugable targets. However, traditional methods for circulating tumor cel identification and isolation often compromise mRNA integrity, due to cell handling such as the cell permeabilization, required for intracellular staining. Here, we present a novel identification and isolation pipeline for single cell transcriptomics of magnetically enriched tumor by combining rVAR2 based identification cells with single-cell magnetic pickup.
Methods
To test our method, we used tumor cells from four lung cancer cell lines spiked into blood. After magnetic enrichment using the CellSearch® Profile Kit, the resulting sample was divided for staining with either rVAR2 or CellSearch reagents. Single cells were isolated using a magnetic needle and analyzed for mRNA integrity using reverse transcription quantitative polymerase chain reaction (RT-qPCR) targeting the GAPDH and EpCAM genes.
Results
The integration of single-cell magnetic pickup enabled precise single-cell separation, while the extracellular rVAR2 staining enables mRNA preservation, collectively facilitating the detection of low-expressed genes at the single-cell level. Compared to CellSearch staining, the rVAR2 staining resulted in our pipeline in a approximately 10 to 100 times higher mRNA yield from single magnetically enriched tumor cells.
Conclusion
This mRNA-friendly method enhances our ability to study tumor heterogeneity at the single-cell level and supports the development of personalized cancer therapies, making it a valuable tool for circulating tumor cel research and clinical applications.
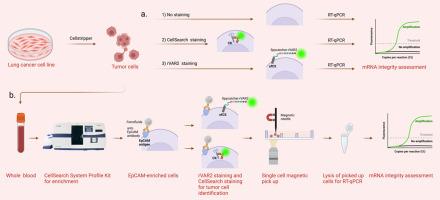
磁富集肿瘤细胞单细胞转录组鉴定及分离途径
循环肿瘤细胞的转录组学分析为癌症转移、异质性和药物靶点的鉴定提供了有价值的见解。然而,传统的循环肿瘤细胞鉴定和分离方法往往会损害mRNA的完整性,因为细胞处理如细胞通透性,需要细胞内染色。在这里,我们提出了一种新的磁性富集肿瘤单细胞转录组学鉴定和分离管道,将基于rVAR2的鉴定细胞与单细胞磁性拾取相结合。方法将四种肺癌细胞系的肿瘤细胞注入血液中进行实验。使用CellSearch®Profile Kit进行磁富集后,将所得样品分开,用rVAR2或CellSearch试剂进行染色。用磁针分离单细胞,用反转录定量聚合酶链反应(RT-qPCR)检测GAPDH和EpCAM基因的mRNA完整性。结果单细胞磁拾取的整合实现了精确的单细胞分离,而细胞外rVAR2染色实现了mRNA的保存,共同促进了单细胞水平低表达基因的检测。与CellSearch染色相比,rVAR2染色使我们的管道从单个磁性富集的肿瘤细胞中获得大约10到100倍的mRNA产量。结论这种mrna友好的方法增强了我们在单细胞水平上研究肿瘤异质性的能力,支持了癌症个性化治疗的发展,使其成为循环肿瘤细胞研究和临床应用的有价值的工具。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


