FungAMR: a comprehensive database for investigating fungal mutations associated with antimicrobial resistance
IF 19.4
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
Abstract
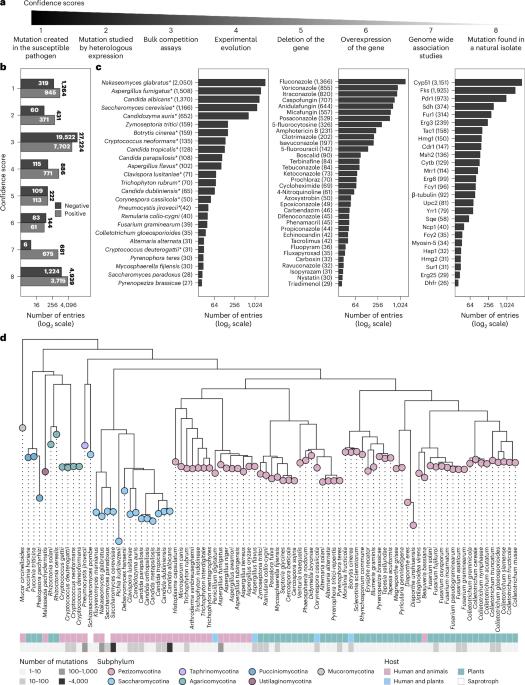
Antimicrobial resistance (AMR) is a global threat, especially in fungal pathogens. To optimize the use of available antifungals, we need rapid detection and monitoring tools that rely on high-quality AMR mutation data. Here we present FungAMR, a resource based on manual curation of 501 published studies on AMR mutations in clinically and agriculturally relevant fungal pathogens resulting in 35,792 entries covering 208 drugs, 246 genes and 95 fungal species. Each entry includes gene, mutation site and drug susceptibility data, with confidence scores indicating the strength of the supporting evidence. Data analysis revealed convergent mechanisms of resistance, indicating some potentially universal resistance mutations and mutations that lead to cross-resistance within and across antifungal classes. We also developed a computational tool, ChroQueTas, that leverages FungAMR to screen fungal genomes for AMR mutations. FungAMR is available as a web-searchable interface within the Comprehensive Antibiotic Resistance Database (CARD). These evolving resources promise to facilitate research on antifungal resistance. An online, manually curated and continually updating database of fungal antimicrobial resistance mutations provides a starting point for further research in fungal AMR.
FungAMR:用于调查与抗菌素耐药性相关的真菌突变的综合数据库
抗微生物药物耐药性(AMR)是一个全球性威胁,特别是在真菌病原体中。为了优化现有抗真菌药物的使用,我们需要依靠高质量AMR突变数据的快速检测和监测工具。FungAMR是一个基于501篇已发表的关于临床和农业相关真菌病原体AMR突变的人工管理研究的资源,共有35,792个条目,涵盖208种药物,246个基因和95种真菌。每个条目包括基因、突变位点和药物敏感性数据,并以置信度评分表示支持证据的强度。数据分析揭示了趋同的耐药机制,表明一些潜在的普遍耐药突变和导致抗真菌类内部和跨抗真菌类交叉耐药的突变。我们还开发了一个计算工具,ChroQueTas,它利用FungAMR筛选真菌基因组中的AMR突变。FungAMR可在综合抗生素耐药性数据库(CARD)中作为网络搜索界面提供。这些不断发展的资源有望促进抗真菌耐药性的研究。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


