Towards a digital twin: Digitization and model-based optimization of the innovative high-gradient magnetic separator
IF 4
Q2 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
Abstract
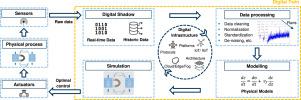
Downstream processing in biotechnology relies on multiple unit operations to achieve high product purity, driving up costs, time, and yield losses. High-Gradient Magnetic Separation (HGMS) offers a promising alternative by consolidating steps and enabling direct target capture from complex media. However, its industrial adoption is hindered by suboptimal performance, limited scalability, and insufficient automation for reproducibility. Furthermore, process efficiency is often not fully realized due to the reliance on fixed operational recipes.
This study presents a digital twin framework for a pilot-scale HGMS system, integrating real-time monitoring, automated control, advanced mechanistic models, and multi-objective optimization using Bayesian algorithms. The framework was validated for robustness, scalable data handling, and predictive control. Key contributions include the development of soft sensors, automated control strategies for improved reproducibility, in-silico optimization of a human Immunoglobulin G (hIgG) capture process — a monoclonal antibody broadly relevant in biopharmaceutical applications — with real-time pH adjustment, and a sensitivity analysis of objective weights, revealing trade-offs between yield, resource use, and processing time. Optimization results indicated a theoretical 4% productivity gain and a 3 percentage point yield improvement, while exposing critical design constraints in the HGMS chamber.
These findings underscore the potential of digital twins to accelerate process optimization and reduce development costs through in-silico experimentation. Future work will focus on refining identified design limitations and extending the framework to optimize process conditions for diverse bioproducts, enhancing scalability and efficiency.
迈向数字孪生:创新高梯度磁选机的数字化与模型优化
生物技术的下游加工依赖于多个单元操作来实现高产品纯度,这增加了成本、时间和产量损失。高梯度磁分离(HGMS)通过巩固步骤和从复杂介质中直接捕获目标,提供了一种有前途的替代方案。然而,它的工业应用受到性能欠佳、可扩展性有限和可重复性自动化不足的阻碍。此外,由于依赖于固定的操作配方,流程效率往往无法完全实现。本研究提出了一个中试规模HGMS系统的数字孪生框架,集成了实时监测、自动控制、先进的机制模型和使用贝叶斯算法的多目标优化。该框架在鲁棒性、可扩展数据处理和预测控制方面得到了验证。主要贡献包括软传感器的开发,提高再现性的自动化控制策略,人类免疫球蛋白G (hIgG)捕获过程的硅片优化-一种在生物制药应用中广泛相关的单克隆抗体-实时pH调节,以及客观权重的敏感性分析,揭示产量,资源利用和处理时间之间的权衡。优化结果表明,理论上生产率提高了4%,产率提高了3个百分点,同时暴露了HGMS腔室的关键设计约束。这些发现强调了数字孪生在通过硅实验加速流程优化和降低开发成本方面的潜力。未来的工作将集中在改进确定的设计限制和扩展框架,以优化各种生物制品的工艺条件,提高可扩展性和效率。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Current Research in Biotechnology
Biochemistry, Genetics and Molecular Biology-Biotechnology
CiteScore
6.70
自引率
3.60%
发文量
50
审稿时长
38 days
期刊介绍:
Current Research in Biotechnology (CRBIOT) is a new primary research, gold open access journal from Elsevier. CRBIOT publishes original papers, reviews, and short communications (including viewpoints and perspectives) resulting from research in biotechnology and biotech-associated disciplines.
Current Research in Biotechnology is a peer-reviewed gold open access (OA) journal and upon acceptance all articles are permanently and freely available. It is a companion to the highly regarded review journal Current Opinion in Biotechnology (2018 CiteScore 8.450) and is part of the Current Opinion and Research (CO+RE) suite of journals. All CO+RE journals leverage the Current Opinion legacy-of editorial excellence, high-impact, and global reach-to ensure they are a widely read resource that is integral to scientists' workflow.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


