A sensitive split luciferase assay for detecting olfactory receptor response to odorants
IF 2.5
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
In this study, olfactory receptors (ORs) and Golf proteins fused with split luciferase were examined to develop a sensitive assay for identifying OR ligands. Although specific luminescence was observed from HEK293T cells expressing both fusion proteins (OR1A1-LgBiT and SmBiT-Mini-Golf) in response to an OR1A1 ligand, its intensity was low. To improve it, a fusion protein (V1N-G4S3-SmBiT-G4S3-Mini-Golf), was created where V1N contained the palmitoylation site necessary for localizing Mini-Golf to the cell membrane. When OR1A1-LgBiT and V1N-G4S3-SmBiT-G4S3-Mini-Golf were co-expressed in HEK293T cells, specific luminescence by ligand stimulation was considerably increased. The cell surface expression of OR1A1-LgBiT was markedly lower than that of OR1A1. Co-expression of receptor transporting protein 1 short form (RTP1S) promoted the cell surface expression of OR1A1-LgBiT more efficiently than OR1A1. Prediction using AlphaFold3 suggested that the N-terminal domain of RTP1S interacts with LgBiT in OR1A1-LgBiT. Co-expression of OR1A1-LgBiT, V1N-G4S3-SmBiT-G4S3-Mini-Golf, and RTP1S in HEK293T cells enhanced the sensitivity for detecting at least some ligands by 1000- to 3000-fold compared to the conventional cAMP assay. These findings suggest that optimizing the assembly of OR1A1-LgBiT and SmBiT-Mini-Golf at the cell membrane is essential for developing a sensitive assay. The OR response-detection system employing split luciferase holds significant potential for investigating ligands of orphan ORs.
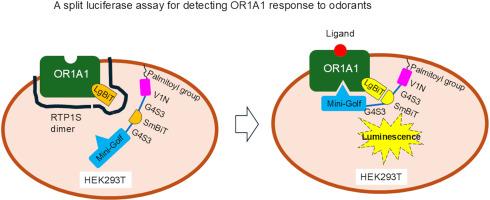
一种灵敏的荧光素酶分裂试验,用于检测嗅觉受体对气味的反应。
在这项研究中,研究嗅觉受体(ORs)和与分裂荧光素酶融合的Golf蛋白,以建立一种识别OR配体的敏感试验。虽然在表达OR1A1- lgbit和SmBiT-Mini-Golf两种融合蛋白的HEK293T细胞中观察到对OR1A1配体的特异性发光,但其强度很低。为了改进它,一个融合蛋白(V1N- g4s3 - smbit - g4s3 -Mini-Golf)被创造出来,其中V1N含有将Mini-Golf定位到细胞膜所必需的棕榈酰化位点。当OR1A1-LgBiT和V1N-G4S3-SmBiT-G4S3-Mini-Golf在HEK293T细胞中共表达时,配体刺激的特异性发光显著增加。OR1A1- lgbit的细胞表面表达量明显低于OR1A1。受体转运蛋白1短链(RTP1S)的共表达比OR1A1更有效地促进OR1A1- lgbit的细胞表面表达。利用AlphaFold3进行预测表明,RTP1S的n端结构域与OR1A1-LgBiT中的LgBiT相互作用。OR1A1-LgBiT、V1N-G4S3-SmBiT-G4S3-Mini-Golf和RTP1S在HEK293T细胞中的共表达,与传统的cAMP检测相比,至少可以将检测某些配体的灵敏度提高1000至3000倍。这些发现表明,优化OR1A1-LgBiT和SmBiT-Mini-Golf的组装对于建立灵敏的检测方法至关重要。采用分裂荧光素酶的OR反应检测系统在研究孤儿OR配体方面具有重要的潜力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical biochemistry
生物-分析化学
CiteScore
5.70
自引率
0.00%
发文量
283
审稿时长
44 days
期刊介绍:
The journal''s title Analytical Biochemistry: Methods in the Biological Sciences declares its broad scope: methods for the basic biological sciences that include biochemistry, molecular genetics, cell biology, proteomics, immunology, bioinformatics and wherever the frontiers of research take the field.
The emphasis is on methods from the strictly analytical to the more preparative that would include novel approaches to protein purification as well as improvements in cell and organ culture. The actual techniques are equally inclusive ranging from aptamers to zymology.
The journal has been particularly active in:
-Analytical techniques for biological molecules-
Aptamer selection and utilization-
Biosensors-
Chromatography-
Cloning, sequencing and mutagenesis-
Electrochemical methods-
Electrophoresis-
Enzyme characterization methods-
Immunological approaches-
Mass spectrometry of proteins and nucleic acids-
Metabolomics-
Nano level techniques-
Optical spectroscopy in all its forms.
The journal is reluctant to include most drug and strictly clinical studies as there are more suitable publication platforms for these types of papers.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


