Analysis of individual patient pathway coordination in a cross-species single-cell kidney atlas
IF 29
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
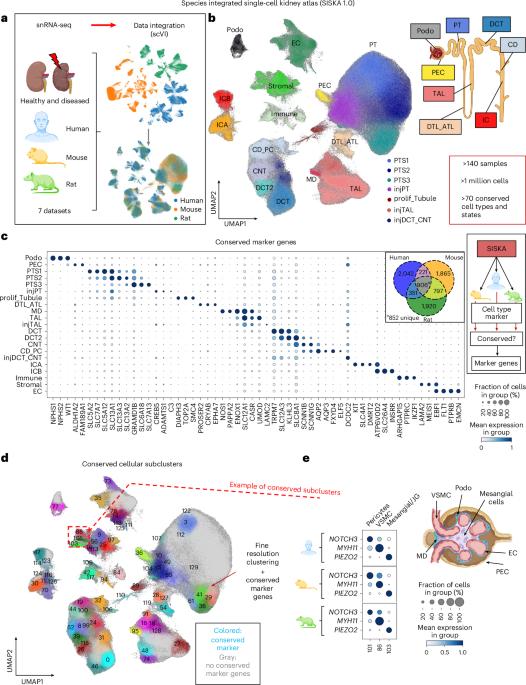
The use of single-cell RNA sequencing in clinical and translational research is limited by the challenge of identifying cell-type-specific, targetable molecular changes in individual patients and cross-species differences. Here we created an integrated single-cell kidney atlas including over 1 million cells from 140 samples, defining more than 70 conserved cell states in human and rodent models. We developed CellSpectra, a computational tool that quantifies changes in gene expression coordination across cellular functions, which we applied to kidney and lung cancer data. This tool powers our patient-level single-cell functional profiling report, which highlights cell-type-specific changes in the coordination of pathway gene expression in individuals. Our cross-species atlas facilitates the selection of a rodent model that closely reflects the cellular and pathway-level signatures observed in patient samples, advancing the application of single-cell methodologies in clinical precision medicine. Finally, using experimental models, we demonstrate how our informatics approach can be applied for the potential selection of suitable therapeutics. CellSpectra identifies functionally relevant changes in cellular pathways when analyzing single samples in comparison to a reference atlas, here applied to rodent models and patient samples of kidney disease.
跨物种单细胞肾图谱中个体患者通路协调分析
单细胞RNA测序在临床和转化研究中的应用受到了在个体患者和跨物种差异中识别细胞类型特异性、可靶向的分子变化的挑战。在这里,我们创建了一个集成的单细胞肾脏图谱,包括来自140个样本的100多万个细胞,在人类和啮齿动物模型中定义了70多种保守的细胞状态。我们开发了CellSpectra,这是一种量化细胞功能中基因表达协调变化的计算工具,我们将其应用于肾脏和肺癌的数据。该工具为我们的患者级单细胞功能分析报告提供了支持,该报告强调了个体中通路基因表达协调中的细胞类型特异性变化。我们的跨物种图谱促进了啮齿动物模型的选择,该模型密切反映了在患者样本中观察到的细胞和途径水平特征,促进了单细胞方法在临床精准医学中的应用。最后,使用实验模型,我们展示了我们的信息学方法如何应用于合适治疗方法的潜在选择。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


