Noncoding rare variant associations with blood traits in 166,740 UK Biobank genomes
IF 29
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
Large biobanks with whole-genome sequencing (WGS) now enable the association of noncoding rare variants with complex human traits. Given that >98% of the genome is available for exploration, the selection of noncoding variants remains a critical yet unresolved challenge in these analyses. Here we leverage knowledge of blood gene regulation and deleteriousness scores to select noncoding variants pertinent for association with blood-related traits. Integrating WGS and 42 blood cell count and biomarker measurements for 166,740 UK Biobank samples, we perform variant collapsing tests, identifying hundreds of gene–trait associations involving noncoding variants. However, we demonstrate that most of these noncoding rare variant associations (1) reproduce associations known from previous studies and (2) are driven by linkage disequilibrium between nearby common and rare variants. This study underscores the prevailing challenges in rare variant analysis and the need for caution when interpreting noncoding rare variant association results. Noncoding rare variant analyses using whole-genome sequencing data from the UK Biobank identify gene–trait associations for 42 blood cell traits but find that most signals are driven by linkage disequilibrium between common and rare variants.

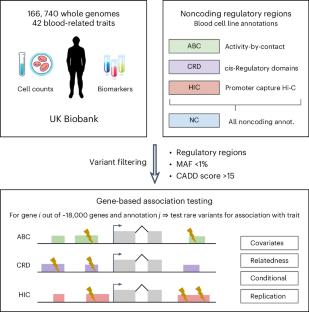
英国生物银行166740个基因组中与血液特征相关的非编码罕见变异
拥有全基因组测序(WGS)的大型生物库现在能够将非编码罕见变异与复杂的人类特征联系起来。鉴于98%的基因组可供探索,非编码变异的选择仍然是这些分析中一个关键但尚未解决的挑战。在这里,我们利用血液基因调控和有害评分的知识来选择与血液相关性状相关的非编码变异。整合WGS和42个血细胞计数以及166740个UK Biobank样本的生物标志物测量,我们进行了变异崩溃测试,确定了数百个涉及非编码变异的基因性状关联。然而,我们证明了这些非编码罕见变异关联中的大多数(1)再现了以前研究中已知的关联,(2)是由附近常见和罕见变异之间的连锁不平衡驱动的。这项研究强调了罕见变异分析中普遍存在的挑战,以及在解释非编码罕见变异关联结果时需要谨慎。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


