Gut bacteria degrade purines via the 2,8-dioxopurine pathway
IF 19.4
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
Abstract
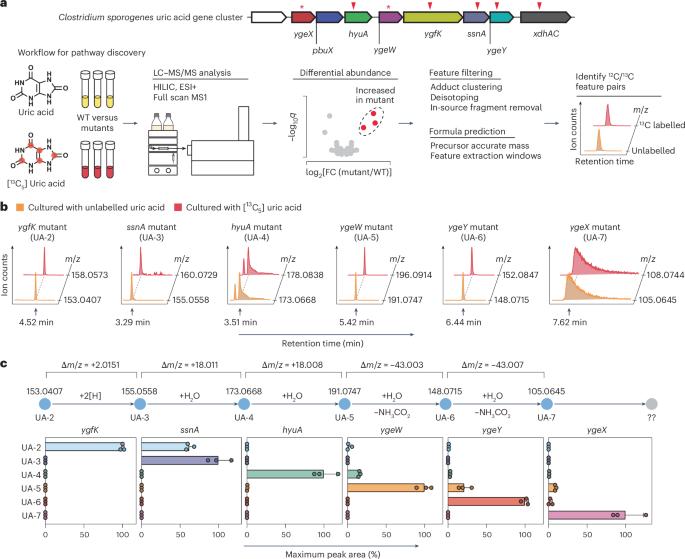
Approximately one-third of urate, which at elevated levels contributes to hyperuricaemia and gout, is excreted into the intestinal tract of healthy individuals where bacteria aid its elimination. However, the molecular details of purine metabolism in the gut microbiome are unclear. Here we uncovered the 2,8-dioxopurine pathway, an anaerobic route for purine degradation in the gut bacteria, Clostridium sporogenes and Escherichia coli. Reconstitution with purified enzymes and mutational analysis combined with isotope tracking and mass spectrometry identified a selenium-dependent enzyme, 2,8-dioxopurine dehydrogenase (DOPDH), and seven additional enzymes that connect purine metabolism to short-chain fatty acid synthesis and ATP generation (measured via luciferase assay). Competition experiments in gnotobiotic mice showed that bacteria harbouring this pathway exhibit a fitness advantage, with wild-type bacteria rapidly outcompeting a DOPDH-deficient strain. Widespread presence of these genes across host-associated microbiomes suggests a host–microbe symbiosis, where host-secreted urate fosters a metabolic niche for bacteria that break it down. These findings could have therapeutic implications for the modification and enhancement of intestinal elimination of urate. Utilizing a selenium-dependent enzyme, gut bacteria degrade uric acid via a previously unrecognized anaerobic pathway which gives them a competitive advantage in the gut.
肠道细菌通过2,8-二氧嘌呤途径降解嘌呤
大约三分之一的尿酸盐会导致高尿酸血症和痛风,它会被排泄到健康个体的肠道中,在那里细菌会帮助消除它。然而,肠道微生物组中嘌呤代谢的分子细节尚不清楚。在这里,我们发现了2,8-二氧嘌呤途径,这是肠道细菌,芽孢梭菌和大肠杆菌中嘌呤降解的厌氧途径。用纯化酶进行重组,结合同位素跟踪和质谱分析,鉴定出一种硒依赖性酶,2,8-二氧嘌呤脱氢酶(DOPDH),以及另外七种将嘌呤代谢与短链脂肪酸合成和ATP生成联系起来的酶(通过荧光素酶测定)。在非生物小鼠中进行的竞争实验表明,拥有这种途径的细菌表现出适应性优势,野生型细菌迅速击败dopdh缺陷菌株。这些基因在宿主相关微生物群中的广泛存在表明存在宿主-微生物共生关系,其中宿主分泌的尿酸为分解它的细菌培养了一个代谢生态位。这些发现可能对改变和增强肠道消除尿酸盐具有治疗意义。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


