The transcriptomics of Hyphantria cunea salivary gland reveals its function in host plant adaptation
IF 3.7
2区 农林科学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Insects' salivary glands and their secretions play a pivotal role in their adaptation to host plants. Hyphantria cunea, a significant pest of both agriculture and forestry, feeds on a variety of host plants, yet the specific functions of its salivary glands in this adaptation process remain largely unclear. In this study, we compared the adaptability of fifth-instar larvae to Populus davidiana × P. bolleana (PDB) and artificial diet (AD). Through transcriptome analysis, 1439 differentially expressed genes (DEGs) were identified in the salivary glands of fifth-instar larvae feeding on PDB and AD. These DEGs include genes encoding various digestive and detoxification enzymes, which are enriched in pathways related to salivary secretion, digestion, and drug metabolism. Compared to larvae fed AD, the majority of digestive and detoxification enzyme genes were upregulated following consumption of PDB. Furthermore, the larvae enhanced the activities of two digestive enzymes (α-amylase and lipase) and four detoxification enzymes (cytochrome P450 monooxygenase, carboxylesterase, glutathione-S-transferase, and UDP-glycosyltransferases) in their salivary glands, thereby digesting the nutrients in the leaves while detoxifying the secondary metabolites contained within them. Silencing of CYP9E2 significantly reduced larval food intake and weight gain, and prolonged larval developmental duration. Taken together, our study identifies the salivary glands of H. cunea larvae as a critical tissue for executing digestive and detoxification functions, enhances understanding of the larval adaptability to host plants via salivary glands, and provides valuable insights for managing H. cunea infestations.
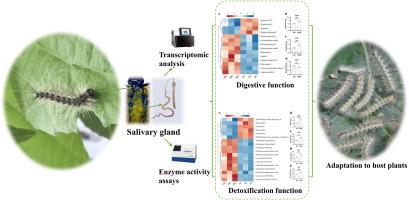
加利福尼亚棘球蚴唾液腺转录组学揭示了其在寄主植物适应中的功能
昆虫的唾液腺及其分泌物在它们适应寄主植物的过程中起着关键作用。美国棘球蚴(Hyphantria cunea)是农业和林业的一种重要害虫,它以多种寄主植物为食,但其唾液腺在这一适应过程中的具体功能仍不清楚。本研究比较了大叶杨(Populus davidiana × P. bolleana, PDB)和人工饲料(artificial diet, AD)对5龄幼虫的适应性。通过转录组分析,在取食PDB和AD的五龄幼虫的唾液腺中鉴定出1439个差异表达基因(DEGs)。这些deg包括编码各种消化和解毒酶的基因,这些基因在唾液分泌、消化和药物代谢的相关途径中富集。与饲喂AD的幼虫相比,食用PDB后,大部分消化酶和解毒酶基因上调。此外,幼虫提高了唾液腺中2种消化酶(α-淀粉酶和脂肪酶)和4种解毒酶(细胞色素P450单加氧酶、羧酯酶、谷胱甘肽- s -转移酶和udp -糖基转移酶)的活性,从而消化叶片中的营养物质,同时对叶片中的次生代谢物进行解毒。CYP9E2的沉默显著降低了幼虫的食物摄取量和体重增加,延长了幼虫的发育时间。综上所述,我们的研究确定了美洲锥虫幼虫的唾液腺是执行消化和解毒功能的关键组织,增强了对幼虫通过唾液腺对寄主植物适应性的认识,并为管理美洲锥虫侵染提供了有价值的见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
7.40
自引率
5.30%
发文量
105
审稿时长
40 days
期刊介绍:
This international journal publishes original contributions and mini-reviews in the fields of insect biochemistry and insect molecular biology. Main areas of interest are neurochemistry, hormone and pheromone biochemistry, enzymes and metabolism, hormone action and gene regulation, gene characterization and structure, pharmacology, immunology and cell and tissue culture. Papers on the biochemistry and molecular biology of other groups of arthropods are published if of general interest to the readership. Technique papers will be considered for publication if they significantly advance the field of insect biochemistry and molecular biology in the opinion of the Editors and Editorial Board.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


