Crystal structures of PAK2 reveal new insights into its autoinhibitory mechanism
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Type I p21-activated kinases (PAK1/2/3) exist in an auto-inhibited form and are stimulated by small G-protein binding and auto-phosphorylation. Previous structural and biochemical studies suggested that PAK1 is a dimer in crystals, and probably in a trans-inhibited conformation in solution. Here, we used multiple independent biochemical and biophysical methods to determine the oligomeric state and autoinhibitory mechanism of PAK2. Crystal structures of the full-length and N-terminal truncated PAK2 reveal the molecular basis underlying the PAK2 autoinhibition. Analytical ultracentrifugation studies show that these proteins have molecular weights that are consistent with monomeric species. The solution-phase structure of the full-length PAK2 by small angle X-ray scattering and computational modeling further shows a compact but elongated molecular shape. These results, taken together with the results of previous studies, demonstrate that in contrast with the most widely accepted model, all three type I PAKs are monomeric in solution and auto-inhibited in cis before activation.
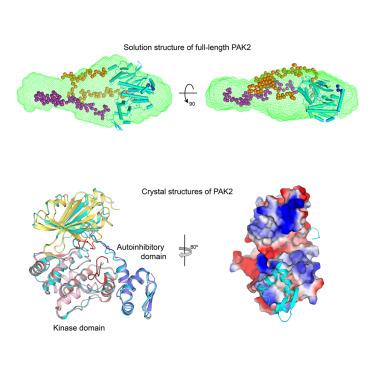
PAK2的晶体结构揭示了其自抑制机制的新见解
I型p21活化激酶(PAK1/2/3)以自抑制形式存在,并通过小g蛋白结合和自磷酸化受到刺激。先前的结构和生化研究表明PAK1在晶体中是二聚体,并且可能在溶液中呈反式抑制构象。在这里,我们使用多种独立的生化和生物物理方法来确定PAK2的寡聚状态和自身抑制机制。全长PAK2和n端截断PAK2的晶体结构揭示了PAK2自抑制的分子基础。分析性超离心研究表明,这些蛋白质具有与单体物种一致的分子量。通过x射线小角散射和计算建模得到全长PAK2的溶液相结构进一步显示出紧凑而细长的分子形状。这些结果与先前的研究结果一起表明,与最广泛接受的模型相反,所有三种I型PAKs在溶液中都是单体的,并且在激活前在cis中被自动抑制。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


