Genome-wide analysis of 3′ untranslated region alternative polyadenylation quantitative trait loci identified a potential novel susceptibility locus for lung cancer in cross-ancestry populations
IF 2.5
3区 生物学
Q2 GENETICS & HEREDITY
引用次数: 0
Abstract
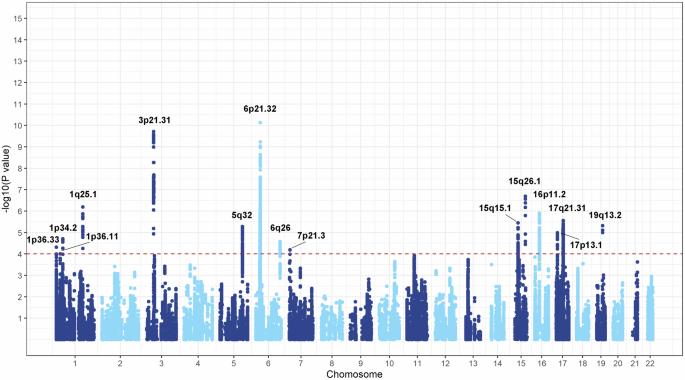
Genome wide association studies (GWASs) have revealed several lung cancer susceptibility loci; however, we still face key issues such as how to identify more ‘high-frequency and inefficient’ variants and decipher the causal variants. Alternative polyadenylation (APA) can shorten or prolong the 3′UTR of mRNA containing cis regulatory elements, which plays an important role in post transcriptional regulation. Specific variants can affect the 3′UTR APA, leading to differences in the risk of lung cancer among individuals carrying different alleles. In this study, a cross-ancestry two-stage case-control design was used to evaluate the associations of 3′UTR APA related variants (3′aQTL SNPs, 3′aSNPs) defined by GTEx in normal lung tissues with the risk of lung cancer based on the genome-wide meta-analysis (GWMA) from FinnGen, UK Biobank and VA Million Veteran Program (MVP), including 28,557 cases and 1,355,961 controls. The promising variants were validated in an independent Chinese population with 1169 lung cancer cases and 1046 controls. Functional annotations of the identified 3′aSNP and related genes were performed based on multiple public databases. Finally, we identified a potential 3′aSNP rs11583258 associated with the risk of lung cancer both in the GWMA [OR = 1.04 (1.02–1.06), P = 1.00 × 10–4] and in the validation stage [OR = 1.08 (1.01–1.16), P = 1.01 × 10–2] at 1p36.11. Function annotation integrating the results of multiple public datasets suggested the variants in this region might affect both the length of the 3′UTR of the UBXN11 transcripts and the expression of UBXN11 to affect the susceptibility of lung cancer.
对3'非翻译区选择性多聚腺苷酸化数量性状位点的全基因组分析发现了一个潜在的新的跨祖先人群肺癌易感位点。
全基因组关联研究(GWASs)揭示了几个肺癌易感位点;然而,我们仍然面临着一些关键问题,比如如何识别更多的“高频和低效”变体,并破译因果变体。选择性聚腺苷化(APA)可缩短或延长含有顺式调控元件的mRNA的3'UTR,在转录后调控中起重要作用。特定的变异可以影响3'UTR APA,导致携带不同等位基因的个体患肺癌的风险存在差异。本研究采用跨血统两阶段病例对照设计,基于FinnGen、UK Biobank和VA Million Veteran Program (MVP)的全基因组荟萃分析(GWMA),评估正常肺组织中由GTEx定义的3'UTR APA相关变异(3'aQTL SNPs、3'aSNPs)与肺癌风险的相关性,包括28,557例和1,355,961例对照。这些有希望的变异在1169例肺癌病例和1046例对照的独立中国人群中得到了验证。基于多个公共数据库对鉴定的3'aSNP及相关基因进行功能注释。最后,我们在GWMA [OR = 1.04 (1.02-1.06), P = 1.00 × 10-4]和验证阶段[OR = 1.08 (1.01-1.16), P = 1.01 × 10-2]和1p36.11中发现了潜在的3'aSNP rs11583258与肺癌风险相关。综合多个公开数据集结果的功能注释表明,该区域的变异可能同时影响UBXN11转录本的3'UTR长度和UBXN11的表达,从而影响肺癌的易感性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Human Genetics
生物-遗传学
CiteScore
7.20
自引率
0.00%
发文量
101
审稿时长
4-8 weeks
期刊介绍:
The Journal of Human Genetics is an international journal publishing articles on human genetics, including medical genetics and human genome analysis. It covers all aspects of human genetics, including molecular genetics, clinical genetics, behavioral genetics, immunogenetics, pharmacogenomics, population genetics, functional genomics, epigenetics, genetic counseling and gene therapy.
Articles on the following areas are especially welcome: genetic factors of monogenic and complex disorders, genome-wide association studies, genetic epidemiology, cancer genetics, personal genomics, genotype-phenotype relationships and genome diversity.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


