Sub-3 Å resolution protein structure determination by single-particle cryo-EM at 100 keV
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Cryoelectron microscopy (cryo-EM) has transformed structural biology by providing high-resolution insights into biological macromolecules. We report sub-3 Å resolution structures using the 100 keV Tundra cryo-TEM, equipped with the Falcon C direct electron detector (DED). This system combines advanced optics, extreme-brightness field emission gun (XFEG), and SP-TWIN lens to enhance coherence and resolution. The semi-automated loader reduced contamination and drift, enabling extended data collection, while the high detective quantum efficiency (DQE) of Falcon C improved signal-to-noise ratio. We validated performance by determining structures of biological samples, including apoferritin (2.1 Å), T20S proteasome (2.7 Å), GABAA receptor (2.8 Å), hemoglobin (5.0 Å), transthyretin (3.5 Å), and AAV9 capsid (2.8 Å), spanning 50 kDa–3.9 MDa. This work highlights the potential of 100 keV transmission electron microscopes (TEMs) to make cryo-EM more accessible. It sets a precedent for using lower voltage TEMs not only for screening, but also for high-resolution protein structure determination.
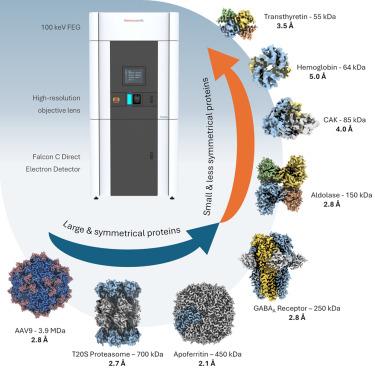
亚-3 Å分辨率蛋白质结构测定,单粒子低温电镜在100 keV
低温电子显微镜(cryo-EM)通过提供对生物大分子的高分辨率见解,改变了结构生物学。我们报告了使用配备猎鹰C直接电子探测器(DED)的100 keV冻土带冷冻透射电镜(Tundra cro - tem)的亚3 Å分辨率结构。该系统结合了先进的光学器件、极亮场发射枪(XFEG)和SP-TWIN透镜,以提高相干性和分辨率。半自动装载机减少了污染和漂移,扩展了数据收集,而猎鹰C的高探测量子效率(DQE)提高了信噪比。我们通过测定生物样品的结构来验证性能,包括载铁蛋白(2.1 Å), T20S蛋白酶体(2.7 Å), GABAA受体(2.8 Å),血红蛋白(5.0 Å),转甲状腺素(3.5 Å)和AAV9衣壳(2.8 Å),跨越50 kDa-3.9 MDa。这项工作强调了100 keV透射电子显微镜(tem)的潜力,使低温电镜更容易获得。它开创了使用低电压tem不仅用于筛选,而且用于高分辨率蛋白质结构测定的先例。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


