The food-associated resistome is shaped by processing and production environments
IF 19.4
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
Abstract
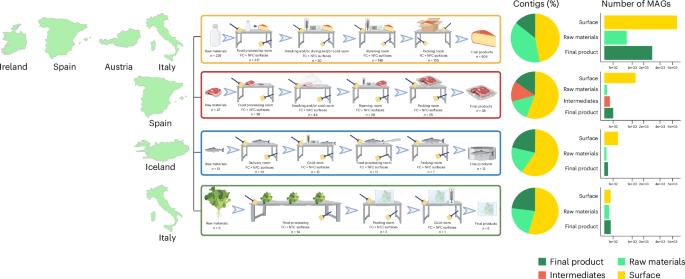
Food production systems may act as transmission routes for antimicrobial-resistant (AMR) bacteria and AMR genes (AMRGs) to humans. However, the food resistome remains poorly characterized. Here 1,780 raw-material (milk, brine, fresh meat and so on), end-product (cheese, fish, meat products and vegetables) and surface (processing, cooling, smoking, ripening and packing rooms) samples from 113 food processing facilities were subjected to whole-metagenome sequencing. Assembly-free analyses demonstrated that >70% of all known AMRGs, including many predicted to confer resistance to critically important antibiotics, circulate throughout food production chains, with those conferring resistance to tetracyclines, β-lactams, aminoglycosides and macrolides being the most abundant overall. An assembly-based analysis highlighted that bacteria from the ESKAPEE group, together with Staphylococcus equorum and Acinetobacter johnsonii, were the main AMRG carriers. Further evaluation demonstrated that ~40% of the AMRGs were associated with mobile genetic elements, mainly plasmids. These findings will help guide the appropriate use of biocides and other antimicrobials in food production settings when designing efficient antimicrobial stewardship policies. Whole-metagenome sequencing of 1,780 raw-material, end-product and surface samples from 113 food processing facilities reveals the occurrence of antimicrobial resistance determinants in foods and their processing environments.

与食物相关的抵抗组受加工和生产环境的影响
粮食生产系统可能是抗微生物药物耐药性(AMR)细菌和抗微生物药物耐药性基因(AMRGs)向人类传播的途径。然而,食物抵抗组的特征仍然很差。在这里,来自113个食品加工设施的1780个原料(牛奶、卤水、鲜肉等)、最终产品(奶酪、鱼、肉制品和蔬菜)和表面(加工、冷却、熏制、成熟和包装室)样本进行了全宏基因组测序。无组装分析表明,70%的已知AMRGs,包括许多被预测会对至关重要的抗生素产生耐药性的AMRGs,在整个食品生产链中流通,其中对四环素类、β-内酰胺类、氨基糖苷类和大环内酯类产生耐药性的AMRGs数量最多。基于组合的分析强调,来自ESKAPEE组的细菌,以及equorum葡萄球菌和johnsonii不动杆菌是AMRG的主要携带者。进一步评价表明,约40%的AMRGs与移动遗传元件相关,主要是质粒。这些发现将有助于指导在制定有效的抗菌素管理政策时在食品生产环境中适当使用杀菌剂和其他抗菌素。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


