Pangenome analysis provides insights into legume evolution and breeding
IF 29
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
Grain legumes hold great promise for advancing sustainable agriculture. Although the evolutionary history of legume species has been investigated, the conserved mechanisms that drive adaptive evolution and govern agronomic improvement remain elusive. Here we present high-quality genome assemblies for nine widely consumed pulses, including common bean, chickpea, pea, lentil, faba bean, pigeon pea, cowpea, mung bean and hyacinth bean. Pangenome analysis reveals the expansion of distinct gene sets in cool-season and warm-season legumes, highlighting the role of gene birth and duplication in the autoregulation of nodulation. Notably, hundreds of genes undergo convergent selection during the evolution of legumes, affecting agronomic traits such as seed weight. In addition, we demonstrate that tandem amplification of transposable elements in gene-depleted regions has a crucial role in driving genome enlargement and the formation of regulatory elements in cool-season legumes. Our results provide insights into the molecular mechanisms underlying the diversification of legumes and represent a valuable resource for facilitating legume breeding. De novo long-read genome assembly for nine legume species and pangenome analysis identify genomic variations associated with environmental adaptation and domestication and highlight the role of transposable elements in genome evolution.
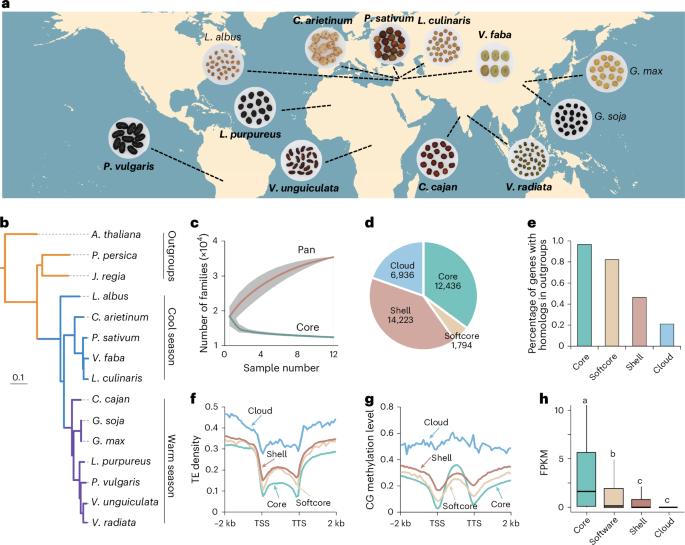

泛基因组分析提供了对豆科植物进化和育种的见解
谷物豆科作物在促进可持续农业方面前景广阔。虽然豆科植物的进化历史已经被研究过,但驱动适应性进化和控制农艺改进的保守机制仍然难以捉摸。在这里,我们展示了9种广泛食用的豆类的高质量基因组组装,包括普通豆、鹰嘴豆、豌豆、扁豆、蚕豆、鸽豆、豇豆、绿豆和风信子豆。泛基因组分析揭示了冷季和暖季豆科植物中不同基因集的扩展,突出了基因出生和复制在结瘤自动调节中的作用。值得注意的是,在豆科植物的进化过程中,数百个基因经历了趋同选择,影响了种子重量等农艺性状。此外,我们证明了基因缺失区域转座因子的串联扩增在驱动冷季豆科植物基因组扩增和调控元件的形成中起着至关重要的作用。本研究结果为揭示豆科植物多样性的分子机制提供了新的思路,为促进豆科植物育种提供了宝贵的资源。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


