Natural variation in SBRR1 shows high potential for sheath blight resistance breeding in rice
IF 29
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
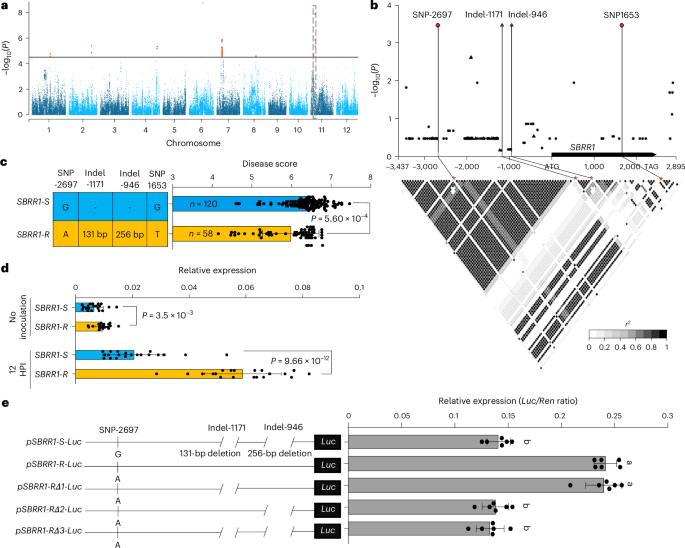
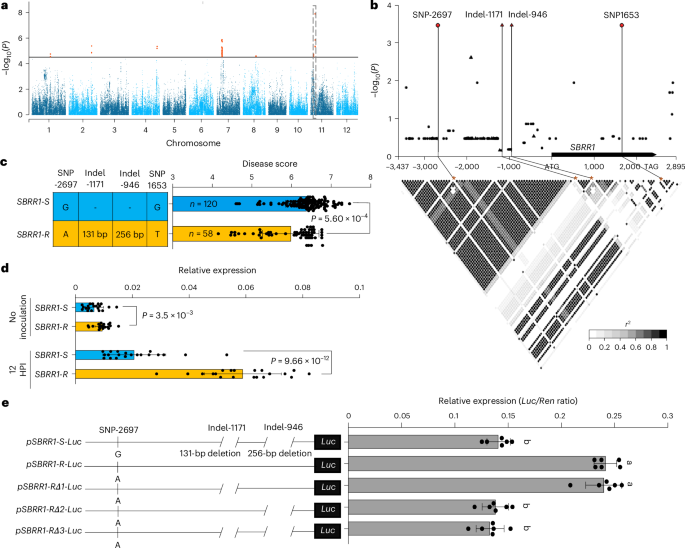
Sheath blight (ShB), caused by necrotrophic fungus Rhizoctonia solani, is one of the most serious rice diseases worldwide. To the best of our knowledge, no genes with high potential for rice ShB resistance breeding have been previously characterized. Here we identify a ShB resistance receptor-like kinase 1 (SBRR1) gene via a genome-wide association study. The SBRR1-R elite allele, containing a 256-bp insertion in its promoter, is preferentially present in indica varieties in geographical regions with highly favorable conditions for ShB development. Introduction of SBRR1-R into a commercial japonica rice variety significantly reduces yield loss under severe ShB disease pressure. Transcription factor bHLH57 specifically binds to the 256-bp sequence and accounts for highly induced expression and stronger resistance of SBRR1-R. Localization of SBRR1 on plasma membrane, aided by SBRR1-interaction-protein 1, and phosphorylation of SBRR1 are required for SBRR1 to rapidly upregulate downstream chitinase genes for resistance. These findings offer mechanistical insights into ShB resistance hidden in natural rice varieties. A genome-wide association study in rice identifies variation in SBRR1 associated with sheath blight resistance. Introducing the SBRR1-R allele into a commercial rice variety reduces yield loss under severe exposure to sheath blight disease.
SBRR1的自然变异在水稻抗纹枯病育种中具有很高的潜力
水稻纹枯病(ShB)是由坏死性真菌枯丝核菌(Rhizoctonia solani)引起的一种严重病害。据我们所知,以前还没有鉴定出水稻稻瘟病抗性育种高潜力的基因。在这里,我们通过全基因组关联研究确定了一个ShB抗性受体样激酶1 (SBRR1)基因。SBRR1-R精英等位基因在启动子上有一个256 bp的插入,该等位基因优先存在于具有高度有利ShB发育条件的地理区域的籼稻品种中。将SBRR1-R引入商业粳稻品种可显著降低在严重ShB病压力下的产量损失。转录因子bHLH57特异性结合256-bp序列,是SBRR1-R高诱导表达和较强抗性的原因。SBRR1在质膜上的定位,在SBRR1相互作用蛋白1的帮助下,以及SBRR1的磷酸化是SBRR1快速上调下游几丁质酶基因产生抗性的必要条件。这些发现提供了对隐藏在天然水稻品种中的ShB抗性的机械见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


