Sliding Window Interaction Grammar (SWING): a generalized interaction language model for peptide and protein interactions
IF 32.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Protein language models embed protein sequences for different tasks. However, these are suboptimal at learning the language of protein interactions. We developed an interaction language model (iLM), Sliding Window Interaction Grammar (SWING) that leverages differences in amino-acid properties to generate an interaction vocabulary. SWING successfully predicted both class I and class II peptide–major histocompatibility complex interactions. Furthermore, the class I SWING model could uniquely cross-predict class II interactions, a complex prediction task not attempted by existing methods. Using human class I and II data, SWING accurately predicted murine class II peptide–major histocompatibility interactions involving risk alleles in systemic lupus erythematosus and type 1 diabetes. SWING accurately predicted how variants can disrupt specific protein–protein interactions, based on sequence information alone. SWING outperformed passive uses of protein language model embeddings, demonstrating the value of the unique iLM architecture. Overall, SWING is a generalizable zero-shot iLM that learns the language of protein–protein interactions. SWING is a versatile interaction language model that can learn the language of peptide and protein interactions.
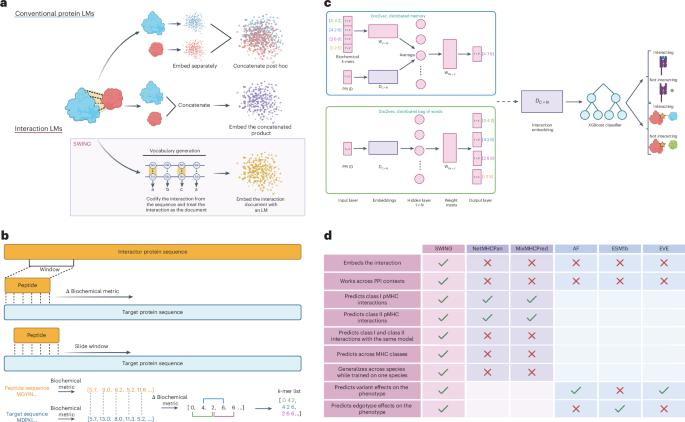
滑动窗口相互作用语法(SWING):肽和蛋白质相互作用的通用相互作用语言模型。
蛋白质语言模型嵌入不同任务的蛋白质序列。然而,这些在学习蛋白质相互作用的语言方面是次优的。我们开发了一个交互语言模型(iLM),滑动窗口交互语法(SWING),它利用氨基酸属性的差异来生成交互词汇表。SWING成功预测了I类和II类肽-主要组织相容性复合体的相互作用。此外,第一类SWING模型可以唯一地交叉预测第二类相互作用,这是现有方法没有尝试过的复杂预测任务。利用人类I类和II类数据,SWING准确预测了涉及系统性红斑狼疮和1型糖尿病风险等位基因的小鼠II类肽-主要组织相容性相互作用。SWING仅基于序列信息就能准确预测变异如何破坏特定蛋白质的相互作用。SWING优于被动使用蛋白质语言模型嵌入,展示了独特的iLM架构的价值。总的来说,SWING是一个通用的零射击iLM,它学习蛋白质-蛋白质相互作用的语言。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


