GeneAgent: self-verification language agent for gene-set analysis using domain databases
IF 32.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Gene-set analysis seeks to identify the biological mechanisms underlying groups of genes with shared functions. Large language models (LLMs) have recently shown promise in generating functional descriptions for input gene sets but may produce factually incorrect statements, commonly referred to as hallucinations in LLMs. Here we present GeneAgent, an LLM-based AI agent for gene-set analysis that reduces hallucinations by autonomously interacting with biological databases to verify its own output. Evaluation of 1,106 gene sets collected from different sources demonstrates that GeneAgent is consistently more accurate than GPT-4 by a significant margin. We further applied GeneAgent to seven novel gene sets derived from mouse B2905 melanoma cell lines. Expert review confirmed that GeneAgent produces more relevant and comprehensive functional descriptions than GPT-4, providing valuable insights into gene functions and expediting knowledge discovery. GeneAgent is a language agent using large language models and self-verification to improve gene-set function annotation.
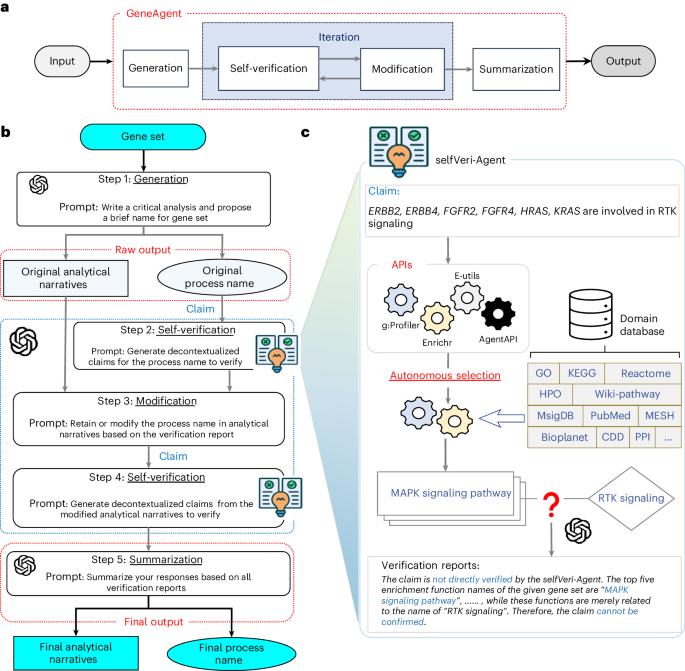
GeneAgent:使用域数据库进行基因集分析的自我验证语言代理。
基因集分析旨在确定具有共享功能的基因群的生物学机制。大型语言模型(llm)最近在为输入基因集生成功能描述方面表现出了希望,但可能会产生事实错误的陈述,在llm中通常被称为幻觉。在这里,我们介绍了GeneAgent,一个基于llm的人工智能代理,用于基因集分析,通过自主与生物数据库交互来验证自己的输出来减少幻觉。对从不同来源收集的1106个基因集的评估表明,GeneAgent始终比GPT-4更准确。我们进一步将GeneAgent应用于来自小鼠B2905黑色素瘤细胞系的七个新基因集。专家评审证实,GeneAgent比GPT-4提供了更相关和全面的功能描述,为基因功能提供了有价值的见解,加快了知识发现。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


