PRDM13 is required for specification of PAX2 lineage inhibitory neurons in the developing cerebellum
IF 2.1
3区 生物学
Q2 DEVELOPMENTAL BIOLOGY
引用次数: 0
Abstract
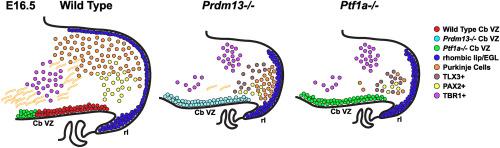
Shared genetic developmental programs in which specific transcription factors affect similar cell fate decisions in distinct tissues are common. In the developing dorsal neural tube and cerebellum, PTF1A is essential for specification of GABAergic inhibitory neurons and suppression of alternative glutamatergic excitatory neuronal fates. Previous studies in the mouse dorsal neural tube identified the transcriptional repressor PRDM13 as a transcriptional target of PTF1A that functions to suppress the alternate cell fates to ensure precision in neuronal cell identity. The presence of PRDM13 in PTF1A + cerebellar progenitors suggests a similar role for PRDM13 in cerebellar neuronal specification. Cerebellar agenesis in humans with missense mutations in PRDM13, and perturbations in cerebellar development in Prdm13 mutant mice and zebrafish, confirm PRDM13 requirement in this tissue. Here we add to these findings showing additional mutant alleles in mouse Prdm13 phenocopy the perturbation in cerebellar cell fates seen with the absence of PTF1A, including loss of PAX2+ interneuron and Purkinje cell inhibitory neuronal lineages, increases in TLX3+ excitatory neuronal lineages, increased apoptosis, and reduced cerebellar size. Additional defects are seen in the placement of TBR1+ cerebellar cells. Thus, using Prdm13 mutant mice, we support conclusions that PRDM13 functions to specify balanced numbers of inhibitory and excitatory neuronal progenitors in the developing cerebellum.
PRDM13是发育中的小脑中PAX2谱系抑制神经元的指定所必需的。
共享的遗传发育程序中,特定的转录因子影响不同组织中相似的细胞命运决定是常见的。在发育中的背神经管和小脑中,PTF1A对于gaba能抑制性神经元的指定和谷氨酸能兴奋性神经元命运的抑制至关重要。先前在小鼠背神经管中的研究发现,转录抑制因子PRDM13是PTF1A的转录靶点,其功能是抑制细胞的交替命运,以确保神经元细胞身份的准确性。PRDM13在PTF1A+小脑祖细胞中的存在表明,PRDM13在小脑神经元分化中具有类似的作用。PRDM13错义突变的人类小脑发育,以及PRDM13突变小鼠和斑马鱼小脑发育的扰动,证实了该组织对PRDM13的需求。在此,我们补充了这些发现,表明小鼠Prdm13中的其他突变等位基因在PTF1A缺失时表现出小脑细胞命运的紊乱,包括PAX2+中间神经元和浦肯野细胞抑制性神经元谱系的缺失,TLX3+兴奋性神经元谱系的增加,凋亡增加和小脑大小的减小。TBR1+小脑细胞的位置也存在其他缺陷。因此,使用Prdm13突变小鼠,我们支持Prdm13功能在发育中的小脑中指定抑制和兴奋性神经元祖细胞数量平衡的结论。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Developmental biology
生物-发育生物学
CiteScore
5.30
自引率
3.70%
发文量
182
审稿时长
1.5 months
期刊介绍:
Developmental Biology (DB) publishes original research on mechanisms of development, differentiation, and growth in animals and plants at the molecular, cellular, genetic and evolutionary levels. Areas of particular emphasis include transcriptional control mechanisms, embryonic patterning, cell-cell interactions, growth factors and signal transduction, and regulatory hierarchies in developing plants and animals.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


