Metagenomic selections reveal diverse antiphage defenses in human and environmental microbiomes
IF 18.7
1区 医学
Q1 MICROBIOLOGY
引用次数: 0
Abstract
To prevent phage infection, bacteria have developed an arsenal of antiphage defenses. Evidence suggests that many examples in nature have not been described. Using plasmid libraries expressing small DNA inserts and functional selections for antiphage defense in Escherichia coli, we identified over 200 putative defenses from 14 bacterial phyla in 9 human and soil microbiomes. Many defenses were unrecognizable based on sequence or predicted structure and thus could only be identified via functional assays. In mechanistic studies, we show that some defenses encode nucleases that distinguish phage DNA via diverse chemical modifications. We also identify outer membrane proteins that prevent phage adsorption and a set of unknown defenses with diverse antiphage profiles and modalities. Most defenses acted against at least two phages, indicating that broadly acting systems are widely distributed. Collectively, these findings highlight the diversity and interoperability of antiphage defense systems.
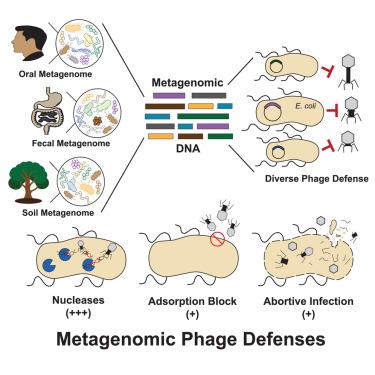
宏基因组选择揭示了人类和环境微生物组中不同的噬菌体防御
为了防止噬菌体感染,细菌已经形成了一系列的抗噬菌体防御系统。有证据表明,自然界中有许多例子没有被描述过。利用表达小片段DNA插入的质粒文库和大肠杆菌抗噬菌体防御的功能选择,我们在9个人类和土壤微生物组中鉴定了来自14个细菌门的200多种推定防御。许多防御无法根据序列或预测结构来识别,因此只能通过功能分析来识别。在机制研究中,我们发现一些防御编码核酸酶,通过不同的化学修饰来区分噬菌体DNA。我们还鉴定了防止噬菌体吸附的外膜蛋白和一系列具有不同噬菌体谱和模式的未知防御。大多数防御措施至少对两种噬菌体起作用,这表明广泛作用的系统分布广泛。总的来说,这些发现突出了抗噬菌体防御系统的多样性和互操作性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cell host & microbe
生物-微生物学
CiteScore
45.10
自引率
1.70%
发文量
201
审稿时长
4-8 weeks
期刊介绍:
Cell Host & Microbe is a scientific journal that was launched in March 2007. The journal aims to provide a platform for scientists to exchange ideas and concepts related to the study of microbes and their interaction with host organisms at a molecular, cellular, and immune level. It publishes novel findings on a wide range of microorganisms including bacteria, fungi, parasites, and viruses. The journal focuses on the interface between the microbe and its host, whether the host is a vertebrate, invertebrate, or plant, and whether the microbe is pathogenic, non-pathogenic, or commensal. The integrated study of microbes and their interactions with each other, their host, and the cellular environment they inhabit is a unifying theme of the journal. The published work in Cell Host & Microbe is expected to be of exceptional significance within its field and also of interest to researchers in other areas. In addition to primary research articles, the journal features expert analysis, commentary, and reviews on current topics of interest in the field.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


