Unlocking expanded flagellin perception through rational receptor engineering
IF 13.6
1区 生物学
Q1 PLANT SCIENCES
引用次数: 0
Abstract
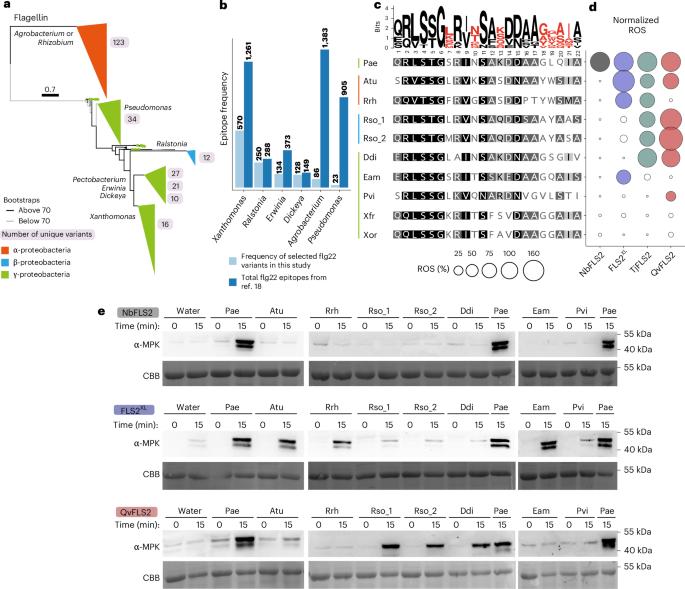
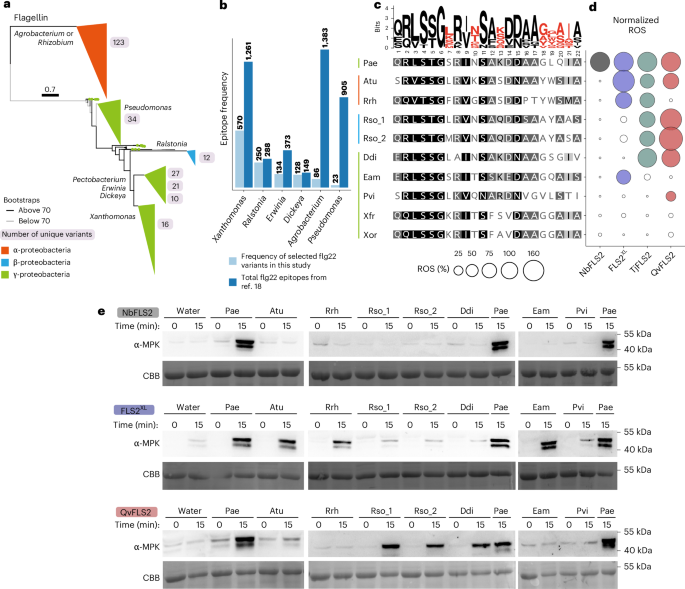
The surface-localized receptor kinase FLS2 detects the flg22 epitope from bacterial flagella. FLS2 is conserved across land plants, but bacterial pathogens exhibit polymorphic flg22 epitopes. Most FLS2 homologues possess narrow perception ranges, but four with expanded perception have been identified. Using diversity analyses, AlphaFold modelling and amino acid properties, key residues enabling expanded recognition were mapped to FLS2’s concave surface, interacting with the co-receptor and polymorphic flg22 residues. Synthetic biology enabled engineering of expanded recognition from QvFLS2 (Quercus variabilis) into a homologue with canonical perception. A similar approach enabled transfer of Agrobacterium perception from FLS2XL (Vitis riparia) into VrFLS2. Evolutionary analyses across three plant orders showed residues under positive selection aligning with those binding the co-receptor and flg22’s C terminus, suggesting more alleles with expanded perception exist. Our experimental data enabled the identification of specific receptor amino acid properties and AlphaFold3 metrics that facilitate predicting FLS2–flg22 recognition. This study provides a framework for rational receptor engineering to enhance pathogen restriction. Receptor kinase FLS2 detects the flg22 epitope of bacterial flagellin. Here the authors identify key residues on FLS2’s concave surface that enable expanded perception of flg22 variants, allowing the engineering of synthetic receptors with enhanced pathogen recognition.
通过理性受体工程解锁扩展的鞭毛蛋白感知
表面定位受体激酶FLS2从细菌鞭毛中检测flg22表位。FLS2在陆地植物中是保守的,但细菌病原体表现出多态性的flg22表位。大多数FLS2同源物具有狭窄的感知范围,但已经确定了四个具有扩展感知的同源物。利用多样性分析、AlphaFold建模和氨基酸特性,将扩展识别的关键残基映射到FLS2的凹表面,并与共受体和多态flg22残基相互作用。合成生物学使扩展识别的工程从QvFLS2 (Quercus variabilis)扩展到具有规范感知的同源物。类似的方法使农杆菌感知从FLS2XL (Vitis riparia)转移到VrFLS2。对三个植物目的进化分析表明,正选择下的残基与结合共受体和flg22的C端一致,表明存在更多具有扩展感知的等位基因。我们的实验数据能够识别特异性受体氨基酸特性和AlphaFold3指标,有助于预测FLS2-flg22的识别。该研究为合理的受体工程提供了框架,以加强病原体的限制。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Plants
PLANT SCIENCES-
CiteScore
25.30
自引率
2.20%
发文量
196
期刊介绍:
Nature Plants is an online-only, monthly journal publishing the best research on plants — from their evolution, development, metabolism and environmental interactions to their societal significance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


