The structure of an ancient genotype–phenotype map shaped the functional evolution of a protein family
IF 13.9
1区 生物学
Q1 ECOLOGY
引用次数: 0
Abstract
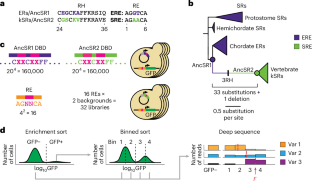
Mutations are more likely to produce some phenotypes than others, but the causal role of these production propensities in the evolution of phenotypic diversity remains unclear. There are two major challenges: it is difficult to separate the effect of the genotype–phenotype (GP) map from that of natural selection when analysing natural diversity, and most extant phenotypes evolved long ago in species whose GP maps cannot be recovered. Here, using two reconstructed ancestral transcription factors that are closely related but differ in function, we created libraries containing all possible amino acid combinations at historically variable sites in the proteins’ DNA binding interface (the genotypes) and measured their capacity to specifically bind DNA elements containing all possible combinations of nucleotides at historically variable sites (the phenotypes). The ancestral GP maps were strongly anisotropic (the distribution of phenotypes encoded by genotypes is highly non-uniform) and heterogeneous (the phenotypes accessible around each genotype vary greatly among genotypes), but the extent and direction of these properties differed substantially between the maps. In both cases, these properties steered evolution towards the lineage-specific phenotypes that evolved during history. Our findings establish that ancient properties of the GP relationship were causal factors in the evolutionary process that produced present-day patterns of functional conservation and diversity. A combination of ancestral reconstruction and deep mutational scanning to examine the genotype–phenotype maps of steroid receptors shows that the properties of ancestral genotype–phenotype maps determine the lineage-specific evolution of DNA specificity.

一个古老的基因型-表型图谱的结构塑造了一个蛋白质家族的功能进化
突变更有可能产生某些表型,但这些生产倾向在表型多样性进化中的因果作用尚不清楚。有两个主要的挑战:在分析自然多样性时,很难将基因型-表现型(GP)图谱的影响与自然选择的影响分开,并且大多数现存的表现型在GP图谱无法恢复的物种中很久以前就进化了。在这里,我们使用两个重建的祖先转录因子,它们密切相关但功能不同,我们创建了包含蛋白质DNA结合界面中历史可变位点(基因型)上所有可能的氨基酸组合的文库,并测量了它们特异性结合包含历史可变位点(表型)上所有可能的核苷酸组合的DNA元件的能力。祖先GP图谱具有很强的各向异性(基因型编码的表型分布高度不均匀)和异质性(每个基因型周围可获得的表型在基因型之间差异很大),但这些特性的程度和方向在图谱之间存在很大差异。在这两种情况下,这些特性都将进化导向了历史上进化的谱系特异性表型。我们的研究结果表明,GP关系的古老属性是进化过程中产生当今功能保护和多样性模式的因果因素。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature ecology & evolution
Agricultural and Biological Sciences-Ecology, Evolution, Behavior and Systematics
CiteScore
22.20
自引率
2.40%
发文量
282
期刊介绍:
Nature Ecology & Evolution is interested in the full spectrum of ecological and evolutionary biology, encompassing approaches at the molecular, organismal, population, community and ecosystem levels, as well as relevant parts of the social sciences. Nature Ecology & Evolution provides a place where all researchers and policymakers interested in all aspects of life's diversity can come together to learn about the most accomplished and significant advances in the field and to discuss topical issues. An online-only monthly journal, our broad scope ensures that the research published reaches the widest possible audience of scientists.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


