Protein–ligand data at scale to support machine learning
IF 51.7
1区 化学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
Abstract
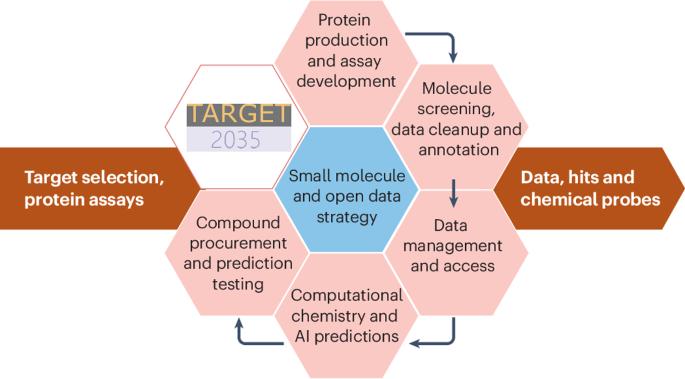
Target 2035 is a global initiative that aims to develop a potent and selective pharmacological modulator, such as a chemical probe, for every human protein by 2035. Here, we describe the Target 2035 roadmap to develop computational methods to improve small-molecule hit discovery, which is a key bottleneck in the discovery of chemical probes. Large, publicly available datasets of high-quality protein–small-molecule binding data will be created using affinity-selection mass spectrometry and DNA-encoded chemical library screening. Positive and negative data will be made openly available, and the machine learning community will be challenged to use these data to build models and predict new, diverse small-molecule binders. Iterative cycles of prediction and testing will lead to improved models and more successful predictions. By 2030, Target 2035 will have identified experimentally verified hits for thousands of human proteins and advanced the development of open-access algorithms capable of predicting hits for proteins for which there are not yet any experimental data. Target 2035 aims to develop a potent and selective pharmacological modulator for every human protein by 2035 with the results made publicly available. This Roadmap article sets out how that will be achieved.
大规模的蛋白质配体数据支持机器学习。
“目标2035”是一项全球倡议,旨在到2035年为每种人类蛋白质开发一种有效的、选择性的药理调节剂,如化学探针。在这里,我们描述了目标2035路线图,以开发计算方法来提高小分子命中发现,这是发现化学探针的关键瓶颈。使用亲和选择质谱法和dna编码化学文库筛选,将创建大型、公开的高质量蛋白质-小分子结合数据集。正面和负面数据将公开提供,机器学习社区将面临挑战,使用这些数据建立模型并预测新的、多样化的小分子粘合剂。预测和测试的迭代循环将导致改进的模型和更成功的预测。到2030年,“2035目标”将确定数千种经过实验验证的人类蛋白质,并推进开放获取算法的发展,这些算法能够预测尚未获得任何实验数据的蛋白质的命中。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature reviews. Chemistry
Chemical Engineering-General Chemical Engineering
CiteScore
52.80
自引率
0.80%
发文量
88
期刊介绍:
Nature Reviews Chemistry is an online-only journal that publishes Reviews, Perspectives, and Comments on various disciplines within chemistry. The Reviews aim to offer balanced and objective analyses of selected topics, providing clear descriptions of relevant scientific literature. The content is designed to be accessible to recent graduates in any chemistry-related discipline while also offering insights for principal investigators and industry-based research scientists. Additionally, Reviews should provide the authors' perspectives on future directions and opinions regarding the major challenges faced by researchers in the field.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


