Regulatory mechanisms of PP2A complex assembly driven by physicochemical differences in A-subunit isoforms
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Protein phosphatase 2A (PP2A) is crucial for regulating cellular pathways, with its holoenzyme assembly affecting enzyme function and substrate selection. The PP2A holoenzyme comprises scaffold A-, regulatory B-, and catalytic C-subunits, each with various isoforms. Here, we examine structural and biochemical characteristics of the A-subunit isoforms (Aα and Aβ) and identify different biophysical properties that may promote distinct PP2A functions. Our molecular dynamics simulations and cryo-EM analyses define structural differences in the isoforms that reside primarily at the N-terminus of the A-subunit where it interfaces with regulatory B-subunits. Kinetic analyses show Aβ has a lower binding affinity in complexes with B56 subunits and exhibits unique aggregative properties as a monomeric protein. These findings suggest that the different physicochemical properties between A-subunit isoforms are key to PP2A holoenzyme assembly and function. We predict that the Aβ serves as a reservoir, ensuring that serine-threonine phosphatase activity is maintained during high regulatory demand.
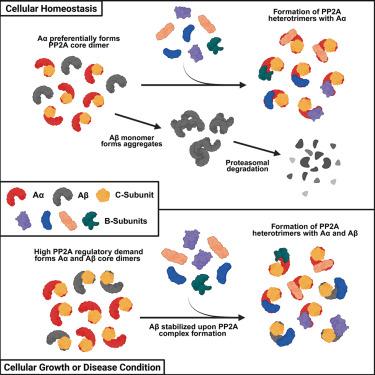
a亚基异构体的物理化学差异驱动PP2A复合体组装的调控机制
蛋白磷酸酶2A (PP2A)对调节细胞通路至关重要,其全酶组装影响酶功能和底物选择。PP2A全酶包括支架A-、调节B-和催化c -亚基,每个亚基都有不同的亚型。在这里,我们研究了a亚基亚型(Aα和Aβ)的结构和生化特征,并确定了可能促进不同PP2A功能的不同生物物理特性。我们的分子动力学模拟和低温电镜分析确定了主要存在于a亚基的n端与调节b亚基接口的同工异构体的结构差异。动力学分析表明,a β与B56亚基复合物具有较低的结合亲和力,并且作为单体蛋白具有独特的聚集特性。这些发现表明,a亚基亚型之间不同的物理化学性质是PP2A全酶组装和功能的关键。我们预测,a β作为一个储存库,确保丝氨酸-苏氨酸磷酸酶活性在高调节需求下维持。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


