Automated removal of corrupted tilts in cryo-electron tomography
IF 5.1
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
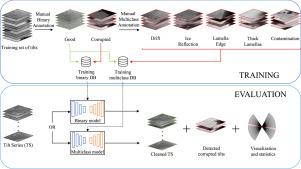
Cryo-electron tomography (cryo-ET) enables the visualization of macromolecular structures in their near-native cellular environment. However, acquired tilt series are often compromised by image corruption due to drift, contamination, and ice reflections. Manually identifying and removing corrupted tilts is subjective and time-consuming, making an automated approach necessary. In this study, we present a deep learning-based method for automatically removing corrupted tilts. We evaluated 13 different neural network architectures, including convolutional neural networks (CNNs) and transformers. Using a dataset of 435 annotated tilt series, we trained models for both binary and multiclass classification of corrupted tilts. We demonstrate the high efficiency and reliability of these automated approaches for removing corrupted tilts in cryo-ET and provide a framework, including models trained on cryo-ET data, that allows users to apply these models directly to their tilt series, improving the quality and consistency of downstream cryo-ET data processing.
冷冻电子断层扫描中损坏倾斜的自动去除
低温电子断层扫描(cryo-ET)使其在接近原生细胞环境中的大分子结构可视化。然而,由于漂移、污染和冰反射,获得的倾斜序列经常受到图像损坏的影响。手动识别和删除损坏的倾斜是主观且耗时的,因此必须采用自动化方法。在这项研究中,我们提出了一种基于深度学习的方法来自动去除损坏的倾斜。我们评估了13种不同的神经网络架构,包括卷积神经网络(cnn)和变压器。使用435个带注释的倾斜序列数据集,我们训练了用于损坏倾斜的二元和多类分类的模型。我们证明了这些自动化方法在去除cryo-ET中损坏的倾斜方面的高效率和可靠性,并提供了一个框架,包括在cryo-ET数据上训练的模型,允许用户将这些模型直接应用于他们的倾斜系列,从而提高下游cryo-ET数据处理的质量和一致性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Structural Biology: X
Biochemistry, Genetics and Molecular Biology-Structural Biology
CiteScore
6.50
自引率
0.00%
发文量
20
审稿时长
62 days
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


