Longitudinal analysis of carbapenem-resistant Escherichia coli in a Southern Chinese hospital (2015–2021): Epidemiology, genetics, and resistance mechanisms
IF 4.6
2区 医学
Q1 INFECTIOUS DISEASES
International Journal of Antimicrobial Agents
Pub Date : 2025-07-19
DOI:10.1016/j.ijantimicag.2025.107577
引用次数: 0
Abstract
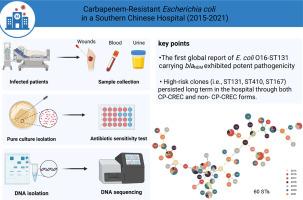
This study conducted a retrospective genomic and epidemiological analysis of Escherichia coli (CREC) in a large general hospital in southern China. From 2015–2021, the characteristics of 183 CREC strainswere determined via minimum inhibitory concentration (MIC) determination and whole-genome sequencing (WGS). The carbapenem resistance mechanisms of carbapenemase-producing carbapenem-resistant E.coli (CP-CREC) and non-carbapenemase-producing carbapenem-resistant E. coli (non-CP-CREC) were investigated through a combination of porin genes (ompC/ompF) assessment and antimicrobial resistance gene detection/copy number quantification. We observed a diverse population structure of CREC, with a total of 60 different sequence types (STs) assigned. A total of 48.6% (89/183) of theisolates were extraintestinal pathogenic E. coli, including the first reported O16-ST131 strains carrying carbapenem resistance genes. Adhesion and invasion experiments revealed that the O16-ST131 CP-CREC strains had strong adhesion and invasion abilities. The majority of CREC strains (90.2%, 165/183) were predicted to have reduced expression of at least one outer membrane porin gene (ompC/ompF). Compared with CP-CREC, non-CP-CREC presented significantly higher copy numbers of extended-spectrum β-lactamase (ESBL) genes (median copy number variants = 3.635 × , P <0.001) and ampC genes (median copy number variants: 2.80 × , P = 0.022). High-risk clones (i.e., ST131, ST410, and ST167) persisted long term in the hospital through both CP-CREC and non-CP-CREC forms. Genetic environment analysis revealed that blaNDM was located in a conserved genetic environment (dsbD - trpF - blaNDM). This work provides a comprehensive description of CREC and offers new insights into CREC surveillance based on WGS in China.
南方某医院耐碳青霉烯类大肠杆菌的纵向分析(2015-2021):流行病学、遗传学和耐药机制
本研究对中国南方某大型综合医院的大肠杆菌(CREC)进行了回顾性基因组学和流行病学分析。2015-2021年,通过最低抑制浓度(MIC)测定和全基因组测序(WGS)测定183株CREC菌株的特征。采用孔蛋白基因(ompC/ompF)评估和耐药基因检测/拷贝数定量相结合的方法,研究产碳青霉烯酶耐碳青霉烯大肠杆菌(CP-CREC)和非产碳青霉烯耐碳青霉烯大肠杆菌(non-CP-CREC)对碳青霉烯的耐药机制。我们观察到CREC的种群结构多样,共有60种不同的序列类型(st)。48.6%(89/183)的分离株为肠外致病性大肠杆菌,包括首次报道的携带碳青霉烯类耐药基因的O16-ST131菌株。粘附和侵袭实验表明,O16-ST131 CP-CREC菌株具有较强的粘附和侵袭能力。预计大多数CREC菌株(90.2%,165/183)至少有一个外膜孔蛋白基因(ompC/ompF)表达降低。与CP-CREC相比,non-CP-CREC呈现明显高于副本数量的extended-spectrumβ内酰胺酶基因(ESBL)(中值拷贝数变异 = 2.27 × ,p = 0.0002)和ampC基因(拷贝数变异中位数:2.905 × ,p = 0.022)。高风险克隆(即ST131、ST410和ST167)通过CP-CREC和非CP-CREC形式在医院长期存在。遗传环境分析表明,blaNDM位于一个保守遗传环境(dsbD - trpF - blaNDM)。这项工作提供了对CREC的全面描述,并为中国基于WGS的CREC监测提供了新的见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
21.60
自引率
0.90%
发文量
176
审稿时长
36 days
期刊介绍:
The International Journal of Antimicrobial Agents is a peer-reviewed publication offering comprehensive and current reference information on the physical, pharmacological, in vitro, and clinical properties of individual antimicrobial agents, covering antiviral, antiparasitic, antibacterial, and antifungal agents. The journal not only communicates new trends and developments through authoritative review articles but also addresses the critical issue of antimicrobial resistance, both in hospital and community settings. Published content includes solicited reviews by leading experts and high-quality original research papers in the specified fields.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


