Genomics-driven discovery of superior alleles and genes for yellow rust resistance in wheat
IF 29
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
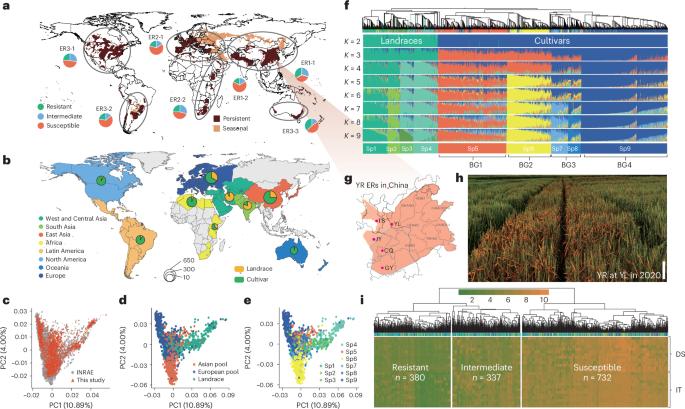
Yellow rust (YR), caused by Puccinia striiformis f. sp. tritici, poses a significant threat to wheat production worldwide. Breeding resistant cultivars is crucial for managing this disease. However, our understanding of the genetic mechanisms underlying YR resistance remains fragmented. To address this, we conducted a comprehensive analysis with variome data from 2,191 wheat accessions worldwide and over 47,000 YR response records across several environments and pathogen races. Through genome-wide association studies, we established a landscape of 431 YR resistance loci, providing a rich resource for resistance gene deployment. Furthermore, we cloned genes corresponding to three resistance loci, namely Yr5x (effective against several P. striiformis f. sp. tritici races), Yr6/Pm5 (conferred resistance to two pathogen species) and YrKB (TaEDR2-B; conferring broad-spectrum rust resistance without yield penalty). These findings offer valuable insights into the genetic basis of YR resistance in wheat and lay the foundation for engineering wheat with durable disease resistance. Genomic analyses of 2,191 global common wheat accessions and over 47,000 yellow rust (YR) response data points across several environments and pathogen races provide a genome-wide landscape of YR resistance genes and effective alleles.

基因组学驱动下小麦抗黄锈优良等位基因和基因的发现
小麦黄锈病(Yellow rust, YR)是由小麦条锈病(Puccinia striiformis f. sp. tritici)引起的一种严重威胁小麦生产的病害。培育抗病品种对控制该病至关重要。然而,我们对YR抗性的遗传机制的理解仍然是碎片化的。为了解决这个问题,我们对全球2191种小麦的各种数据进行了全面分析,并对几种环境和病原体种的47,000多条YR反应记录进行了分析。通过全基因组关联研究,我们建立了431个YR抗性位点的图谱,为抗性基因的配置提供了丰富的资源。此外,我们克隆了三个抗性位点对应的基因,分别是Yr5x(对几种小麦条纹状单胞疟原虫有效)、Yr6/Pm5(对两种病原菌有效)和YrKB (TaEDR2-B;赋予广谱防锈性而不损失产量)。这些发现为小麦抗病的遗传基础提供了有价值的见解,为小麦的持久抗病工程奠定了基础。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


