Molecular representation learning via multimodal fusion and decoupling
IF 15.5
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
Recent years have seen growing attention on self-supervised learning in drug molecule research and discovery. Additionally, a series of methods have emerged that leverage both 2D and 3D structures for molecular representation learning. However, these methods focus only on the modal consistency between 2D and 3D molecular structure relying on molecule-level or atom-level alignment while ignoring modal complementarity. In this paper, we propose a multimodal fusion-then-decoupling self-supervised molecular representation learning method named MolMFD. First, we use a unified encoder to fuse 2D and 3D molecular structural information by incorporating atomic relative distances from both topological and geometric views. Then, we design a learnable noise injection strategy to decouple modality-specific representations, which are subsequently input into separate decoders to predict the structural information of each corresponding modality. Notably, we minimize mutual information to extract the 2D and 3D modality-specific characteristics, considering modality complementarity to enrich the fused molecular representations. We provide a theoretical analysis of the optimization issues and the overlooked complementarity problems in existing 2D and 3D multimodal molecular pre-training methods. Extensive molecular prediction experiments validate the effectiveness and superiority of our proposed MolMFD.
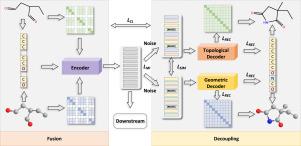
基于多模态融合和解耦的分子表征学习
近年来,自监督学习在药物分子研究和发现中的应用受到越来越多的关注。此外,已经出现了一系列利用二维和三维结构进行分子表征学习的方法。然而,这些方法只关注二维和三维分子结构之间的模态一致性,依赖于分子水平或原子水平的排列,而忽略了模态互补性。本文提出了一种多模态融合-解耦自监督分子表示学习方法MolMFD。首先,我们使用一个统一的编码器融合二维和三维分子结构信息,结合原子相对距离从拓扑和几何视图。然后,我们设计了一个可学习的噪声注入策略来解耦特定于模态的表示,这些表示随后被输入到单独的解码器中,以预测每个相应模态的结构信息。值得注意的是,我们最小化互信息以提取2D和3D模态特定特征,并考虑模态互补性以丰富融合分子表征。我们对现有二维和三维多模态分子预训练方法中的优化问题和被忽视的互补性问题进行了理论分析。大量的分子预测实验验证了我们所提出的MolMFD的有效性和优越性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Information Fusion
工程技术-计算机:理论方法
CiteScore
33.20
自引率
4.30%
发文量
161
审稿时长
7.9 months
期刊介绍:
Information Fusion serves as a central platform for showcasing advancements in multi-sensor, multi-source, multi-process information fusion, fostering collaboration among diverse disciplines driving its progress. It is the leading outlet for sharing research and development in this field, focusing on architectures, algorithms, and applications. Papers dealing with fundamental theoretical analyses as well as those demonstrating their application to real-world problems will be welcome.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


