A structure-guided approach to predict MHC-I restriction of T cell receptors for public antigens
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
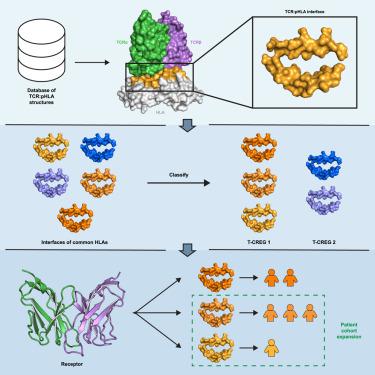
Peptides presented by major histocompatibility complex class I (MHC-I) proteins provide biomarkers for therapeutic targeting using T cell receptors (TCRs), TCR-mimicking antibodies (TMAs), or other engineered protein binders. Despite the extreme sequence diversity of the human leukocyte antigen (HLA, the human MHC), a given TCR or TMA is restricted to recognize epitopic peptides in the context of a limited set of different HLA alleles. Here, guided by our analysis of 98 TCR:pHLA complex structures, we identify TCR contact residues and classify 148 common HLA alleles into T cell cross-reactivity groups (T-CREGs) on the basis of their presented surface features. Insights from our work have actionable value for predicting MHC-I restriction of TCRs, guiding therapeutic expansion of existing TCR-based approaches and informing the selection of peptide targets for the development of new therapeutics.
一种结构导向的方法来预测MHC-I限制T细胞受体的公共抗原
由主要组织相容性复合体I类(MHC-I)蛋白呈递的多肽为使用T细胞受体(tcr)、tcr模拟抗体(tma)或其他工程蛋白结合物进行治疗靶向提供了生物标志物。尽管人类白细胞抗原(HLA,人类MHC)具有极端的序列多样性,但给定的TCR或TMA在有限的不同HLA等位基因背景下只能识别表位肽。在我们对98个TCR:pHLA复合物结构的分析的指导下,我们确定了TCR接触残基,并根据其表面特征将148个常见HLA等位基因分类为T细胞交叉反应基团(T- cregs)。我们的工作见解对于预测tcr的MHC-I限制,指导现有基于tcr的方法的治疗扩展以及为开发新疗法提供肽靶点的选择具有可操作的价值。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


