Jpx RNA controls Xist induction through spatial reorganization of the mouse X-inactivation center
IF 8.7
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
Known to regulate chromosome looping on a genome-wide scale, the noncoding Jpx RNA was originally shown to control X chromosome counting and induce Xist expression during X chromosome inactivation (XCI). Not fully understood is how Jpx upregulates Xist in coordination with Tsix downregulation in cis. Here, by integrating epigenomic data and polymer modeling in a mouse embryonic stem cell model, we demonstrate that Jpx controls architectural and transcriptional dynamics within anti- and pro-XCI zones of the X-inactivation center. Distinct topological changes occur on the future active X (Xa) and inactive X (Xi) chromosomes. Jpx binds the enhancer of Tsix on the future Xi and alters loop formation to favor Xist induction, coordinately releasing CTCF from Tsix and Xist in cis on the future Xi. Thus, by controlling a dynamic rewiring of functional loops, Jpx flips a transcriptional switch to control mutually exclusive Tsix and Xist expression in cis.
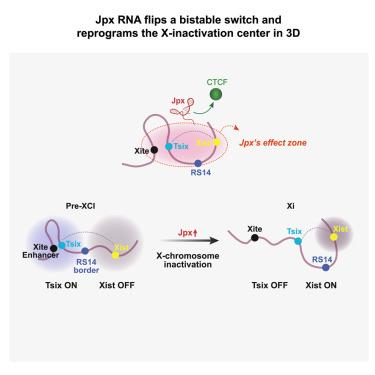
Jpx RNA通过小鼠x -失活中心的空间重组控制Xist的诱导
已知在全基因组范围内调节染色体环,非编码的Jpx RNA最初被证明在X染色体失活(XCI)期间控制X染色体计数并诱导Xist表达。目前还不完全清楚Jpx是如何在cis中上调Xist并协同下调t6的。在这里,通过整合小鼠胚胎干细胞模型的表观基因组数据和聚合物建模,我们证明Jpx控制x -失活中心的抗和促xci区域的结构和转录动力学。在未来的活性X (Xa)和非活性X (Xi)染色体上发生明显的拓扑变化。Jpx在未来Xi上结合Tsix的增强子,改变环的形成有利于Xist的诱导,在未来Xi上协调地从Tsix和Xist释放CTCF。因此,通过控制功能环的动态重新布线,Jpx翻转转录开关来控制cis中互斥的Tsix和Xist表达。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Developmental cell
生物-发育生物学
CiteScore
18.90
自引率
1.70%
发文量
203
审稿时长
3-6 weeks
期刊介绍:
Developmental Cell, established in 2001, is a comprehensive journal that explores a wide range of topics in cell and developmental biology. Our publication encompasses work across various disciplines within biology, with a particular emphasis on investigating the intersections between cell biology, developmental biology, and other related fields. Our primary objective is to present research conducted through a cell biological perspective, addressing the essential mechanisms governing cell function, cellular interactions, and responses to the environment. Moreover, we focus on understanding the collective behavior of cells, culminating in the formation of tissues, organs, and whole organisms, while also investigating the consequences of any malfunctions in these intricate processes.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


