DNA hypomethylation traits define human regulatory T cells in cutaneous tissue and identify their blood recirculating counterparts
IF 27.6
1区 医学
Q1 IMMUNOLOGY
引用次数: 0
Abstract
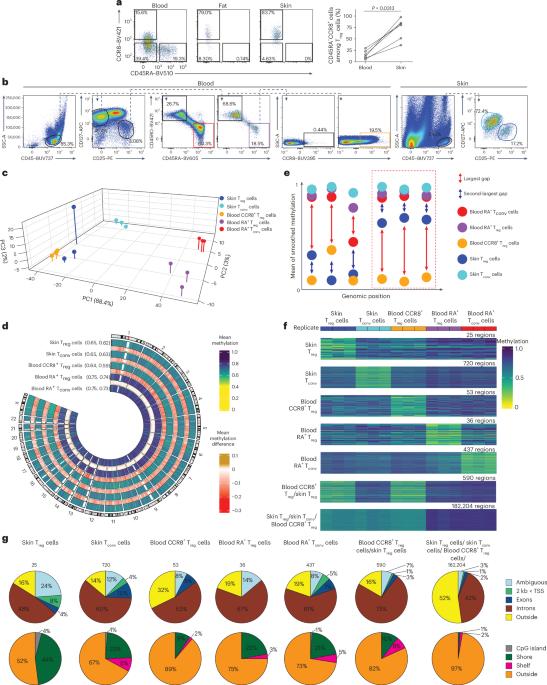
CD4+ regulatory T (Treg) cells in tissues play crucial immunoregulatory and regenerative roles. Despite their importance, the epigenetics and differentiation of human tissue Treg cells are incompletely understood. Here, we performed genome-wide DNA methylation analysis of human Treg cells from skin and blood and integrated these data into a multiomic framework, including chromatin accessibility and gene expression. This analysis identified programs that governed the tissue adaptation of skin Treg cells. We found that subfamilies of transposable elements represented a major constituent of the hypomethylated landscape in tissue Treg cells. Based on T cell antigen receptor sequence and DNA hypomethylation homologies, our data indicate that blood CCR8+ Treg cells contain recirculating human skin Treg cells. Conversely, differences in chromatin accessibility and gene expression suggest a certain reversal of the tissue adaptation program during recirculation. Our findings provide insights into the biology of human tissue Treg cells, which may help harness these cells for therapeutic purposes. Feuerer and colleagues use multiomic analyses to identify DNA methylation, chromatin accessibility and gene expression programs that govern the tissue adaptation of human skin Treg cells.

DNA低甲基化特征定义了皮肤组织中的人类调节性T细胞,并识别了它们的血液循环对应体
CD4+调节性T (Treg)细胞在组织中发挥重要的免疫调节和再生作用。尽管它们很重要,但人类组织Treg细胞的表观遗传学和分化尚未完全了解。在这里,我们对来自皮肤和血液的人类Treg细胞进行了全基因组DNA甲基化分析,并将这些数据整合到多组学框架中,包括染色质可及性和基因表达。该分析确定了控制皮肤Treg细胞组织适应性的程序。我们发现转座因子亚家族代表了组织Treg细胞低甲基化景观的主要组成部分。基于T细胞抗原受体序列和DNA低甲基化同源性,我们的数据表明血液CCR8+ Treg细胞含有循环人皮肤Treg细胞。相反,染色质可及性和基因表达的差异表明在再循环过程中组织适应程序的某种逆转。我们的发现提供了对人体组织Treg细胞生物学的见解,这可能有助于利用这些细胞用于治疗目的。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Immunology
医学-免疫学
CiteScore
40.00
自引率
2.30%
发文量
248
审稿时长
4-8 weeks
期刊介绍:
Nature Immunology is a monthly journal that publishes the highest quality research in all areas of immunology. The editorial decisions are made by a team of full-time professional editors. The journal prioritizes work that provides translational and/or fundamental insight into the workings of the immune system. It covers a wide range of topics including innate immunity and inflammation, development, immune receptors, signaling and apoptosis, antigen presentation, gene regulation and recombination, cellular and systemic immunity, vaccines, immune tolerance, autoimmunity, tumor immunology, and microbial immunopathology. In addition to publishing significant original research, Nature Immunology also includes comments, News and Views, research highlights, matters arising from readers, and reviews of the literature. The journal serves as a major conduit of top-quality information for the immunology community.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


