NMR insights on multidomain proteins: the case of the SARS-CoV-2 nucleoprotein
IF 8.2
2区 化学
Q2 CHEMISTRY, PHYSICAL
Progress in Nuclear Magnetic Resonance Spectroscopy
Pub Date : 2025-07-07
DOI:10.1016/j.pnmrs.2025.101577
引用次数: 0
Abstract
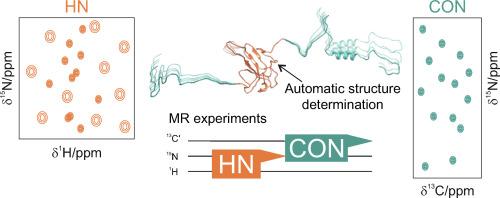
Studying multidomain proteins, especially those combining well-folded domains with intrinsically disordered regions (IDRs), requires specific Nuclear Magnetic Resonance (NMR) techniques to address their structural complexity. To illustrate this, we focus here on the nucleocapsid protein from SARS-CoV-2, which includes both structured and disordered regions. We applied a suite of NMR methods, combining ARTINA software for automatic assignment and structure modelling with multi-receiver experiments that simultaneously capture signals from different nuclear spins, increasing both data quality and acquisition efficiency. Studies of signal temperature-dependence, heteronuclear relaxation and secondary structure propensity (SSP) analysis, as well as experiments employing either 1H or 13C detection to achieve simultaneous snapshots of globular and disordered regions, were used to analyse both the isolated N-terminal domain (NTD) and a construct (NTR) comprising the NTD and two flanking highly disordered regions (IDR1, IDR2). This comprehensive approach allowed us to characterize the NTD's structure and to evaluate how the IDRs affect the overall conformation and dynamics, as well as the interaction with RNA. The findings underscore the importance of applying such a combination of tailored NMR techniques for effectively studying multidomain proteins with heterogeneous structural and dynamic properties.
多结构域蛋白的核磁共振洞察:以SARS-CoV-2核蛋白为例
研究多结构域蛋白质,特别是那些结合良好折叠结构域和内在无序区(IDRs)的蛋白质,需要特定的核磁共振(NMR)技术来解决其结构复杂性。为了说明这一点,我们将重点放在SARS-CoV 2的核衣壳蛋白上,它包括结构区和无序区。我们应用了一套核磁共振方法,结合ARTINA软件进行自动分配和结构建模,以及多接收器实验,同时捕获来自不同核自旋的信号,提高了数据质量和采集效率。研究人员利用信号温度依赖性、异核弛豫和二级结构倾向(SSP)分析,以及利用1H或13C检测实现球状区和无序区同时快照的实验,分析了孤立的n端结构域(NTD)和由NTD和两个侧高无序区(IDR1, IDR2)组成的结构体(NTR)。这种综合方法使我们能够表征NTD的结构,并评估idr如何影响整体构象和动力学,以及与RNA的相互作用。这些发现强调了应用这种定制核磁共振技术的组合来有效研究具有异质结构和动态特性的多结构域蛋白质的重要性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
14.30
自引率
8.20%
发文量
12
审稿时长
62 days
期刊介绍:
Progress in Nuclear Magnetic Resonance Spectroscopy publishes review papers describing research related to the theory and application of NMR spectroscopy. This technique is widely applied in chemistry, physics, biochemistry and materials science, and also in many areas of biology and medicine. The journal publishes review articles covering applications in all of these and in related subjects, as well as in-depth treatments of the fundamental theory of and instrumental developments in NMR spectroscopy.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


