Kaempferol drives genotype-specific microbiota Bacillaceae to enhance nitrogen acquisition in rapeseed
IF 13
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
Introduction
Host genotype is a key driver in shaping plant microbiome in response to dynamic changes in soil nitrogen (N) availability. However, the effects of rapeseed (Brassica napus) genotypes with different N use efficiency characteristics on microbiome assembly, as well as the underlying plant–microbe interaction mechanisms, remain poorly understood.Objectives
This study aims to: (1) assess microbial assembly differences between N-use efficient and inefficient genotypes; (2) identify specific microbiota associated with plant N acquisition; and (3) reveal the molecular mechanisms driving plant–microbe interactions.Methods
We conducted comparative microbiome profiling of N-use efficient and inefficient genotypes, followed by functional validation of microbial roles in plant N uptake. Multi-omics approaches, including RNA-seq and metabolomics, were used to uncover the regulatory interactions between the host and rhizosphere microbiota.Results
The N-use efficient genotype constructed more diverse root-associated microbes than the inefficient genotype, with Bacillaceae emerging as the most enriched taxon. A representative isolate, Bacillus sp. 41S2 from the N-use efficient genotype, markedly enhanced root biomass and N uptake in the N-use inefficient genotype, as confirmed by 15N tracer assays. RNA-Seq analysis further demonstrated that genes involved in jasmonic acid and ethylene signaling pathways were upregulated in strain 41S2-inoculated plants, likely contributing to enhanced root development. Metabolomic profiling identified kaempferol, a flavonol with the highest fold-change between genotypes, as a key root exudate promoting the growth and biofilm formation of strain 41S2. Furthermore, the fls1 mutant (deficient in kaempferol biosynthesis) failed to recruit Bacillaceae, confirming the role of kaempferol in mediating genotype-specific microbial enrichment.Conclusions
Our findings reveal a novel, microbe-dependent N acquisition pathway in N-use efficient rapeseed genotypes, driven by kaempferol-mediated recruitment of Bacillaceae. This work highlights the potential of host genotype and metabolite signaling to shape beneficial microbiota for improved nutrient efficiency and sustainable crop production.
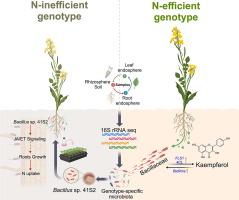
山奈酚驱动基因型特异性菌群杆菌科促进油菜氮素获取
寄主基因型是影响植物微生物组形成以响应土壤氮有效性动态变化的关键驱动因素。然而,具有不同氮素利用效率特征的油菜基因型对微生物组组装的影响以及潜在的植物-微生物相互作用机制尚不清楚。本研究旨在:(1)评估氮利用高效和低效基因型之间的微生物组装差异;(2)确定与植物氮获取相关的特定微生物群;(3)揭示植物与微生物相互作用的分子机制。方法对氮利用高效基因型和氮利用低效基因型进行微生物组分析,并对微生物在植物氮吸收中的作用进行功能验证。多组学方法,包括RNA-seq和代谢组学,被用来揭示宿主和根际微生物群之间的调节相互作用。结果氮利用高效基因型比低效基因型构建了更多的根相关微生物,其中芽孢杆菌科是最丰富的分类群。氮利用高效基因型的代表性分离物芽孢杆菌41S2显著提高了氮利用低效基因型的根生物量和氮吸吸量,15N示踪试验证实了这一点。RNA-Seq分析进一步表明,与茉莉酸和乙烯信号通路相关的基因在菌株41s2接种的植株中表达上调,可能有助于促进根系发育。代谢组学分析发现,山奈酚是促进菌株41S2生长和生物膜形成的关键根分泌物,是基因型之间倍数变化最大的黄酮醇。此外,fls1突变体(缺乏山奈酚的生物合成)未能招募杆菌科,证实了山奈酚在介导基因型特异性微生物富集中的作用。结论研究结果揭示了山奈酚介导的芽孢杆菌科基因型在油菜氮素高效利用中存在一种新的微生物依赖的氮素获取途径。这项工作强调了宿主基因型和代谢物信号传导在塑造有益微生物群以提高养分效率和可持续作物生产方面的潜力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Advanced Research
Multidisciplinary-Multidisciplinary
CiteScore
21.60
自引率
0.90%
发文量
280
审稿时长
12 weeks
期刊介绍:
Journal of Advanced Research (J. Adv. Res.) is an applied/natural sciences, peer-reviewed journal that focuses on interdisciplinary research. The journal aims to contribute to applied research and knowledge worldwide through the publication of original and high-quality research articles in the fields of Medicine, Pharmaceutical Sciences, Dentistry, Physical Therapy, Veterinary Medicine, and Basic and Biological Sciences.
The following abstracting and indexing services cover the Journal of Advanced Research: PubMed/Medline, Essential Science Indicators, Web of Science, Scopus, PubMed Central, PubMed, Science Citation Index Expanded, Directory of Open Access Journals (DOAJ), and INSPEC.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


