A haplotype-resolved pangenome of the barley wild relative Hordeum bulbosum
IF 48.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
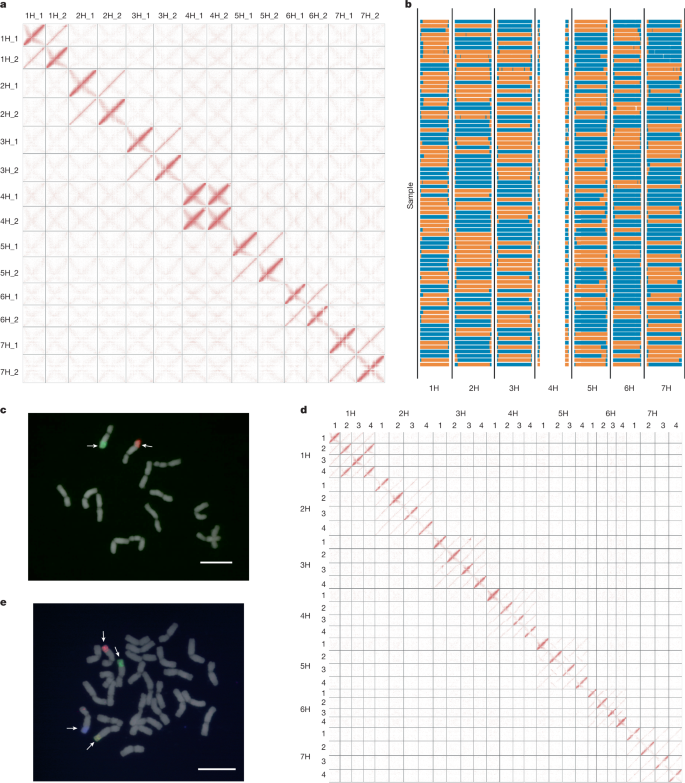
Wild plants can contribute valuable genes to their domesticated relatives1. Fertility barriers and a lack of genomic resources have hindered the effective use of crop–wild introgressions. Decades of research into barley’s closest wild relative, Hordeum bulbosum, a grass native to the Mediterranean basin and Western Asia, have yet to manifest themselves in the release of a cultivar bearing alien genes2. Here we construct a pangenome of bulbous barley comprising 10 phased genome sequence assemblies amounting to 32 distinct haplotypes. Autotetraploid cytotypes, among which the donors of resistance-conferring introgressions are found, arose at least twice, and are connected among each other and to diploid forms through gene flow. The differential amplification of transposable elements after barley and H. bulbosum diverged from each other is responsible for genome size differences between them. We illustrate the translational value of our resource by mapping non-host resistance to a viral pathogen to a structurally diverse multigene cluster that has been implicated in diverse immune responses in wheat and barley. A study describes the assembly and analysis of a haplotype-resolved pangenome of bulbous barley with the potential to improve domesticated barley and illustrates its use in evolutionary research and trait mapping.

大麦野生亲缘种球芽大麦的单倍型解析泛基因组
野生植物可以把有价值的基因提供给它们驯化的亲戚。生育障碍和缺乏基因组资源阻碍了作物-野生基因渗入的有效利用。对大麦最近的野生亲缘种——一种原产于地中海盆地和西亚的草——的研究已经进行了几十年,但还没有发现一种携带外来基因的栽培品种。在这里,我们构建了球茎大麦的全基因组,包括10个阶段的基因组序列,共计32个不同的单倍型。同源四倍体细胞型,其中发现了抗性基因渗入的供体,至少出现了两次,并且通过基因流相互连接并与二倍体形式连接。转座因子的扩增差异是造成大麦和球芽草基因组大小差异的主要原因。我们通过将非宿主对病毒病原体的抗性映射到一个结构多样的多基因簇来说明我们的资源的转化价值,该多基因簇与小麦和大麦的多种免疫反应有关。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature
综合性期刊-综合性期刊
CiteScore
90.00
自引率
1.20%
发文量
3652
审稿时长
3 months
期刊介绍:
Nature is a prestigious international journal that publishes peer-reviewed research in various scientific and technological fields. The selection of articles is based on criteria such as originality, importance, interdisciplinary relevance, timeliness, accessibility, elegance, and surprising conclusions. In addition to showcasing significant scientific advances, Nature delivers rapid, authoritative, insightful news, and interpretation of current and upcoming trends impacting science, scientists, and the broader public. The journal serves a dual purpose: firstly, to promptly share noteworthy scientific advances and foster discussions among scientists, and secondly, to ensure the swift dissemination of scientific results globally, emphasizing their significance for knowledge, culture, and daily life.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


