Redistribution of super-enhancers promotes malignancy in human hepatocellular carcinoma
IF 13
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
Introduction
Super-enhancers (SEs) are defined as the regulatory region where intensive transcriptional cofactors bind. Dysregulation of SEs is related to multiple diseases, however, its role in hepatocellular carcinoma (HCC) remains elusive.Objectives
This work aimed to reveal the dysregulation of SEs in HCC and the therapeutic potential for HCC treatment.Methods
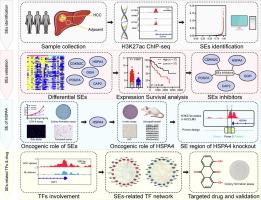
Fifteen HCC and twelve paracancerous samples underwent chromatin immunoprecipitation (ChIP) sequencing targeting H3K27ac, and subsequently the SEs were identified by the Rank Ordering of Super-Enhancers algorithm. Differential SEs featured by tumor or paracancerous tissues were identified, and cross-referenced with the differential expression genes and prognosis-related genes in 2 independent public or in-house HCC cohorts. The SE region of HSPA4 was deleted in the genome of HCCLM3 cell by CRISPR-Cas9, named HSPA4-SE-KO cells. The potential druggable transcriptional factors were identified by CRCmapper, GeneMANIA and Drug Gene Interaction Database (DGID).Results
Five targets, including CDKN2C, HSPA4, GGH, PDGFA, and CAP2, were identified as HCC-gain SEs with oncogenic potential, which were further validated experimentally by SE inhibitors and ChIP targeting H3K27ac and BRD4. Cell proliferation and migration assays further confirmed that silencing of these HCC-gain SEs significantly suppressed the malignant phenotype of HCC cell lines. HSPA4 appeared strongest oncogenic functions among these targets, which was further verified by HCC mouse xenograft models and clinical sample investigation. Moreover, HSPA4-SE-KO cells obtained significantly suppressed HSPA4 expression and retarded tumorigenic capability. Finally, dysregulation of transcriptional factors engaged in the oncogenic role of SEs, and Danthron that targeting RXRA were identified from DGID for HCC treatment.Conclusion
The dysregulated SE landscape of HCC promoted the malignancy phenotype by the upregulation of oncogenes, and SE-regulatory network might be potential drug targets for HCC treatment. Our study deepened the insight of epigenetic dysregulation in HCC, offering the groundwork for SEs as potential therapeutic targets of HCC treatment.
超增强子的重新分配促进人类肝细胞癌的恶性
超级增强子(se)被定义为密集转录辅因子结合的调控区域。SEs的失调与多种疾病有关,但其在肝细胞癌(HCC)中的作用尚不清楚。目的揭示SEs在HCC中的失调及其治疗潜力。方法对15例HCC和12例癌旁肿瘤样本进行靶向H3K27ac的染色质免疫沉淀(ChIP)测序,并通过super - enhancer Rank Ordering算法对se进行鉴定。鉴定出以肿瘤或癌旁组织为特征的差异SEs,并与2个独立的公开或内部HCC队列的差异表达基因和预后相关基因进行交叉对照。通过CRISPR-Cas9将HCCLM3细胞基因组中HSPA4的SE区删除,命名为HSPA4-SE- ko细胞。通过CRCmapper、GeneMANIA和药物基因相互作用数据库(Drug Gene Interaction Database, DGID)对潜在的可药物转录因子进行鉴定。结果CDKN2C、HSPA4、GGH、PDGFA和CAP2等5个靶点被鉴定为具有致癌潜力的hcc -增益SE,并通过SE抑制剂和靶向H3K27ac和BRD4的ChIP进一步实验验证。细胞增殖和迁移实验进一步证实,沉默这些HCC获得性SEs可显著抑制HCC细胞系的恶性表型。HSPA4在这些靶点中表现出最强的致癌功能,这在肝癌小鼠异种移植模型和临床样本调查中得到进一步证实。此外,HSPA4- se - ko细胞显著抑制了HSPA4的表达,并延缓了其致瘤能力。最后,从DGID中鉴定出参与SEs致癌作用的转录因子失调,以及靶向RXRA的丹thron用于HCC治疗。结论肝细胞癌SE格局失调通过上调癌基因促进恶性表型,SE调控网络可能是肝细胞癌治疗的潜在药物靶点。我们的研究加深了对HCC中表观遗传失调的认识,为SEs作为HCC治疗的潜在治疗靶点提供了基础。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Advanced Research
Multidisciplinary-Multidisciplinary
CiteScore
21.60
自引率
0.90%
发文量
280
审稿时长
12 weeks
期刊介绍:
Journal of Advanced Research (J. Adv. Res.) is an applied/natural sciences, peer-reviewed journal that focuses on interdisciplinary research. The journal aims to contribute to applied research and knowledge worldwide through the publication of original and high-quality research articles in the fields of Medicine, Pharmaceutical Sciences, Dentistry, Physical Therapy, Veterinary Medicine, and Basic and Biological Sciences.
The following abstracting and indexing services cover the Journal of Advanced Research: PubMed/Medline, Essential Science Indicators, Web of Science, Scopus, PubMed Central, PubMed, Science Citation Index Expanded, Directory of Open Access Journals (DOAJ), and INSPEC.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


