Single-cell Micro-C profiles 3D genome structures at high resolution and characterizes multi-enhancer hubs
IF 29
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
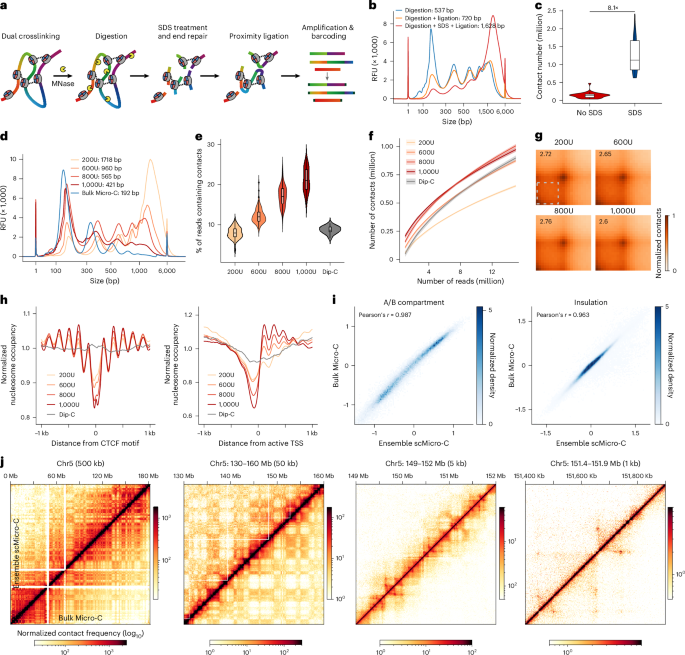
In animal genomes, regulatory DNA elements called enhancers govern precise spatiotemporal gene expression patterns in specific cell types. However, the spatial organization of enhancers within the nucleus to regulate target genes remains poorly understood. Here we report single-cell Micro-C (scMicro-C), a micrococcal nuclease-based three-dimensional (3D) genome mapping technique with an improved spatial resolution of 5 kb, and identified a specialized 3D enhancer structure termed ‘promoter–enhancer stripes (PESs)’, connecting a gene’s promoter to multiple enhancers. PES are formed by cohesin-mediated loop extrusion, which potentially brings multiple enhancers to the promoter. Further, we observed the prevalence of multi-enhancer hubs on genes with PES within single-cell 3D genome structures, wherein multiple enhancers form a spatial cluster in association with the gene promoter. Through its improved resolution, scMicro-C elucidates how enhancers are spatially coordinated to control genes. scMicro-C is a new method that provides high-resolution maps of the 3D genome. scMicro-C identifies structures called ‘promoter stripes’, which link a gene promoter to multiple downstream enhancers.

单细胞Micro-C在高分辨率下描述三维基因组结构,并具有多增强子枢纽的特征
在动物基因组中,被称为增强子的调控DNA元件控制着特定细胞类型中精确的时空基因表达模式。然而,增强子在细胞核内调节靶基因的空间组织仍然知之甚少。在这里,我们报告了单细胞Micro-C (scMicro-C),一种基于微球菌核酸酶的三维(3D)基因组定位技术,其空间分辨率提高到5kb,并确定了一种称为“启动子-增强子条纹(PESs)”的特殊3D增强子结构,将基因的启动子连接到多个增强子。聚醚砜是由内聚素介导的环挤压形成的,这可能会给启动子带来多种增强剂。此外,我们还观察到,在单细胞3D基因组结构中,PES基因上普遍存在多增强子枢纽,其中多个增强子与基因启动子形成空间集群。通过其提高的分辨率,scMicro-C阐明了增强子如何在空间上协调以控制基因。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


