Proteomics profiles of bovine and goat milk fat globule membrane revealed differences on the components and potential functions
Abstract
Background
Goat milk shares a similar nutrient composition with bovine milk and is often used as a substitute in milk product processing. However, goat milk may exhibit distinct biological properties due to compositional differences in its milk fat globule membrane (MFGM), which remain underexplored.
Aim(s)
This study aimed to compare the MFGM proteins of Chinese Holstein bovine and Guanzhong dairy goat milk to elucidate their proteomic profiles and potential nutritional implications.
Methods
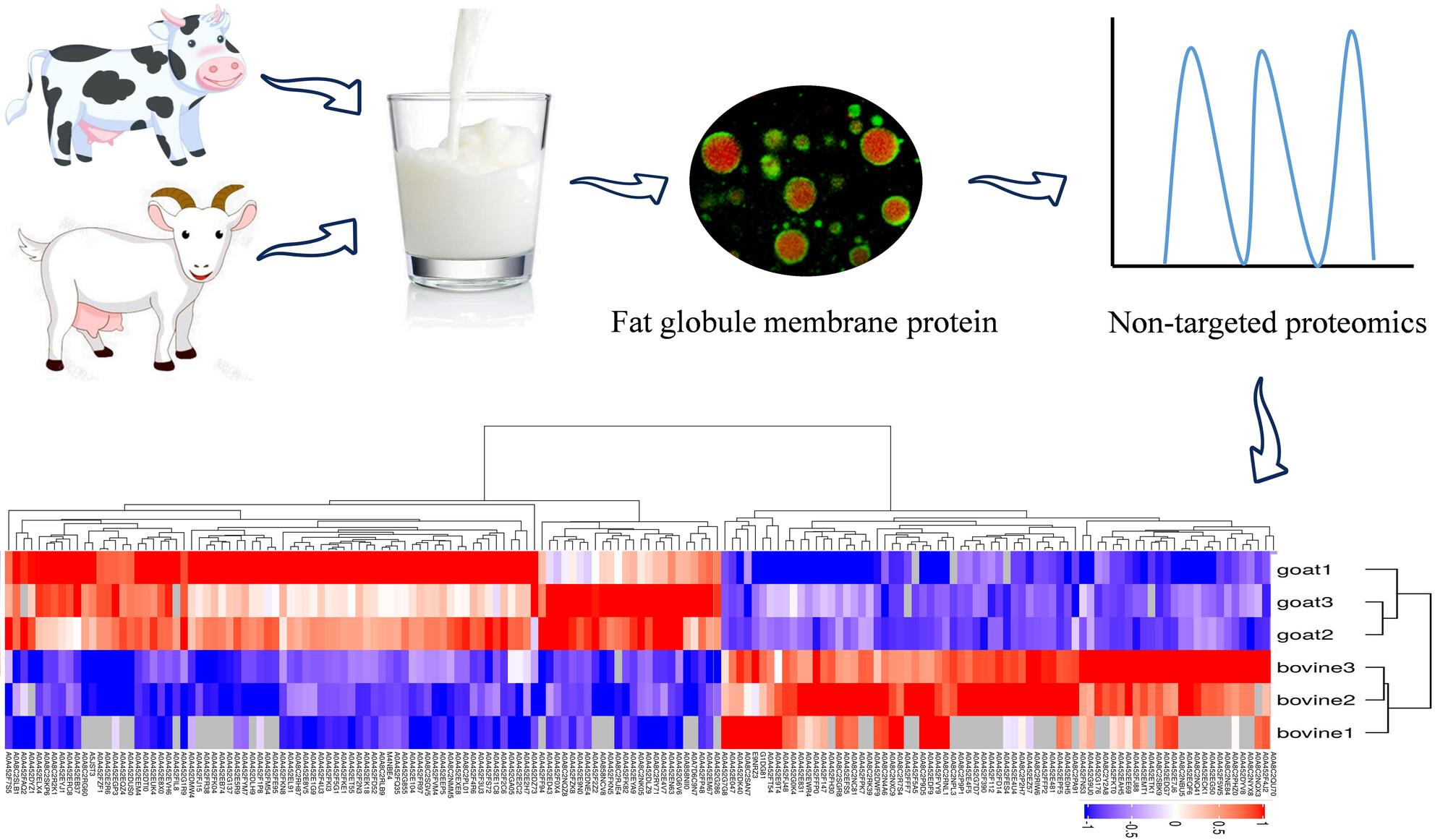
MFGMs were isolated using phosphate-buffered saline aqueous extraction, and their structural integrity was verified using confocal laser scanning microscopy (CLSM). A label-free quantitative proteomics approach was employed to characterise MFGM protein profiles. Functional annotations were performed using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses.
Major findings
CLSM revealed that goat milk MFGMs had weaker structural stability than bovine MFGMs. Quantitative proteomics identified 1414 proteins in bovine MFGM and 2060 in goat MFGM, highlighting significant interspecies divergence in key protein components (P < 0.05). Notably, bovine MFGM demonstrated higher epidermal growth factor expression (P < 0.05), whereas goat MFGM had elevated xanthine oxidoreductase levels (P < 0.05). Differential expression analysis identified 169 signature proteins (e.g. vascular noninflammatory molecule 2, fibrinogen α-chain, and peptidoglycan recognition protein) that distinguished the two milk species. GO analysis revealed that MFGM proteins primarily differed in cellular and metabolic processes. KEGG pathway enrichment analysis further revealed significant differences in pyrimidine metabolism, drug metabolism-cytochrome P450 and glutathione metabolism between bovine and goat MFGMs.
Scientific or industrial implications
This comparative analysis demonstrated that goat MFGM contains unique bioactive proteins and has distinct nutritional properties. The identified biomarkers provide novel targets for dairy authentication and offer new insights into the selection of milk matrices for dairy product processing.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


