Modulation of SARS-CoV-2 spike binding to ACE2 through conformational selection
IF 34.9
1区 材料科学
Q1 MATERIALS SCIENCE, MULTIDISCIPLINARY
引用次数: 0
Abstract
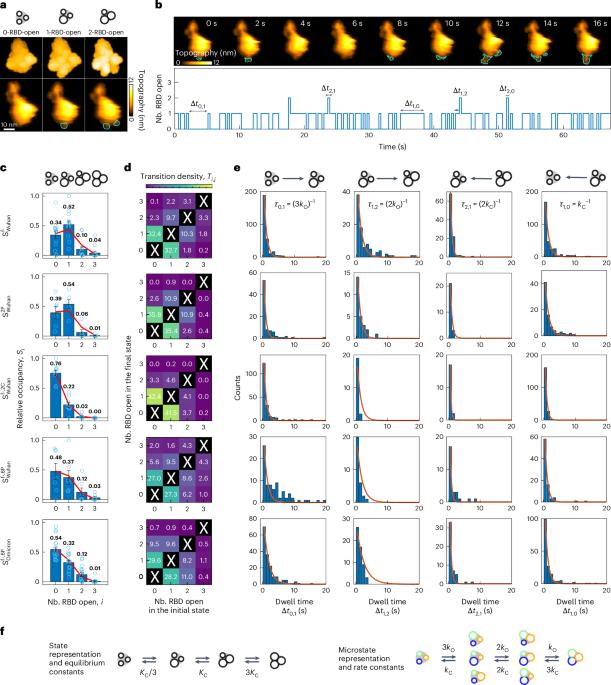
The first step of SARS-CoV-2 infection involves the interaction between the viral trimeric spike protein (S) and the host angiotensin-converting enzyme 2 (ACE2). The receptor-binding domain (RBD) of S adopts two conformations: open and closed, respectively accessible and inaccessible to ACE2. Although these changes surely affect ACE2 binding, a quantitative description of the underlying mechanisms has remained elusive. Here we visualize RBD opening and closing using high-speed atomic force microscopy, gaining access to the corresponding transition rates. We also probe the S/ACE2 interaction at the ensemble level with biolayer interferometry and at the single-molecule level with atomic force microscopy and magnetic tweezers, evidencing that RBD dynamics hinder ACE2 binding but have no effect on unbinding. The resulting modulation is quantitatively predicted by a conformational selection model in which each S protomer behaves independently. Our work thus reveals a molecular mechanism by which RBD accessibility and binding strength can be tuned separately, providing hints to better understand the joint evolution of immune evasion and infectivity. This work uses single-molecule techniques to show that conformational changes of the spike protein of SARS-CoV-2 modulate binding to its human receptor. The study provides key insights into the molecular mechanisms of virus infection and immune evasion.

通过构象选择调节SARS-CoV-2刺突与ACE2的结合
SARS-CoV-2感染的第一步涉及病毒三聚体刺突蛋白(S)与宿主血管紧张素转换酶2 (ACE2)之间的相互作用。S的受体结合域(receptor-binding domain, RBD)有开放和封闭两种构象,分别对ACE2可及和不可及。尽管这些变化肯定会影响ACE2的结合,但对其潜在机制的定量描述仍然难以捉摸。在这里,我们使用高速原子力显微镜观察RBD的打开和关闭,从而获得相应的转变速率。我们还利用生物层干涉法和原子力显微镜和磁镊子在单分子水平上探测了S/ACE2的相互作用,证明RBD动力学阻碍了ACE2的结合,但对解除结合没有影响。由此产生的调制是定量预测的构象选择模型,其中每个S原聚体的行为独立。因此,我们的工作揭示了RBD可达性和结合强度可以分别调节的分子机制,为更好地理解免疫逃避和传染性的联合进化提供了线索。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature nanotechnology
工程技术-材料科学:综合
CiteScore
59.70
自引率
0.80%
发文量
196
审稿时长
4-8 weeks
期刊介绍:
Nature Nanotechnology is a prestigious journal that publishes high-quality papers in various areas of nanoscience and nanotechnology. The journal focuses on the design, characterization, and production of structures, devices, and systems that manipulate and control materials at atomic, molecular, and macromolecular scales. It encompasses both bottom-up and top-down approaches, as well as their combinations.
Furthermore, Nature Nanotechnology fosters the exchange of ideas among researchers from diverse disciplines such as chemistry, physics, material science, biomedical research, engineering, and more. It promotes collaboration at the forefront of this multidisciplinary field. The journal covers a wide range of topics, from fundamental research in physics, chemistry, and biology, including computational work and simulations, to the development of innovative devices and technologies for various industrial sectors such as information technology, medicine, manufacturing, high-performance materials, energy, and environmental technologies. It includes coverage of organic, inorganic, and hybrid materials.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


