cfDNAFE: Comprehensively extracting multi-omics features of cell-free DNA for noninvasive diagnosis
IF 4.3
3区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
The tissues-of-origin of circulating cell-free DNA (cfDNA) holds great promise for non-invasive diagnosing cancers, monitoring allograft rejection, and prenatal testing. Many features for inferring the tissues-of-origin of cfDNAs are being revealed from different angles, including genetics, epigenetics, and fragmentomics, with whole-genome sequencing (WGS) and whole-genome bisulfite sequencing (WGBS) data of cfDNA. However, it lacks integrative toolkits for automatically extracting the revealed features from the WGS and WGBS data of cfDNA samples. Here, we propose cfDNAFE, a comprehensive and easy-to-use python package for extracting multi-omics features from the aligned cfDNA sequencing data. It covers three aspects: cfDNA genetic features, cfDNA methylation features, and cfDNA fragmentation features, including 13 types of feature profiles. The genetic features include substitution mutations, mutation signatures and copy number variations. The methylation features are the proportions of methylated fragments, unmethylated fragments, and mixed methylated fragments on cell-type-specific markers. The fragmentation features related to the fragment sizes, end/breakpoint motifs, and nucleosome positions are also integrated. To verify the functions of cfDNAFE, we perform analysis on the WGS/WGBS data of cfDNA samples based on the feature profiles extracted by cfDNAFE. The comparison between the cfDNA samples of hepatocellular carcinoma (HCC) patients and normal controls suggests HCC cfDNA samples exhibit significant difference in fragment size related features and breakpoint/end motif patterns, and obtain significant higher OCF values in the liver-specific open regions than the health controls. Conclusively, cfDNAFE is a most comprehensive toolkit which covers the most features for inferring the tissues-of-origin of cfDNAs in existing studies up to date. It will facilitate researchers to build machine learning models for auxiliary diagnosis based on these features. Availability and implementation: https://github.com/Cuiwanxin1998/cfDNAFE.
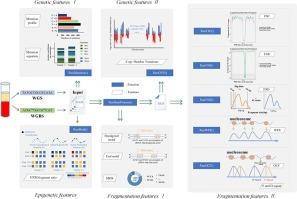
cfDNAFE:综合提取游离DNA的多组学特征,用于无创诊断。
循环无细胞DNA (cfDNA)的起源组织在非侵入性诊断癌症、监测同种异体移植排斥反应和产前检测方面具有很大的前景。利用cfDNA的全基因组测序(WGS)和亚硫酸盐全基因组测序(WGBS)数据,从遗传学、表观遗传学和片段组学等不同角度揭示了推断cfDNA起源组织的许多特征。然而,目前还缺乏从cfDNA样本的WGS和WGBS数据中自动提取揭示特征的集成工具。在这里,我们提出cfDNAFE,一个全面且易于使用的python包,用于从校准的cfDNA测序数据中提取多组学特征。涵盖cfDNA遗传特征、cfDNA甲基化特征和cfDNA片段化特征三个方面,包括13种类型的特征谱。遗传特征包括替代突变、突变特征和拷贝数变异。甲基化特征是甲基化片段、非甲基化片段和混合甲基化片段在细胞类型特异性标记上的比例。还集成了与片段大小、末端/断点基序和核小体位置相关的片段特征。为了验证cfDNAFE的功能,我们基于cfDNAFE提取的特征轮廓对cfDNA样本的WGS/WGBS数据进行了分析。肝细胞癌(HCC)患者cfDNA样本与正常对照的比较表明,HCC cfDNA样本在片段大小相关特征和断点/末端基序模式上存在显著差异,并且在肝脏特异性开放区域获得显著高于健康对照的OCF值。最后,cfDNAFE是一个最全面的工具包,涵盖了迄今为止现有研究中推断cfdna起源组织的大多数特征。这将有助于研究人员基于这些特征构建辅助诊断的机器学习模型。可用性和实现:https://github.com/Cuiwanxin1998/cfDNAFE。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Methods
生物-生化研究方法
CiteScore
9.80
自引率
2.10%
发文量
222
审稿时长
11.3 weeks
期刊介绍:
Methods focuses on rapidly developing techniques in the experimental biological and medical sciences.
Each topical issue, organized by a guest editor who is an expert in the area covered, consists solely of invited quality articles by specialist authors, many of them reviews. Issues are devoted to specific technical approaches with emphasis on clear detailed descriptions of protocols that allow them to be reproduced easily. The background information provided enables researchers to understand the principles underlying the methods; other helpful sections include comparisons of alternative methods giving the advantages and disadvantages of particular methods, guidance on avoiding potential pitfalls, and suggestions for troubleshooting.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


