Comparative structural insights of X protein across species and the “lost” BH3-like domain that may explain the absence of hepatocellular carcinoma in birds
IF 2.6
4区 医学
Q3 INFECTIOUS DISEASES
引用次数: 0
Abstract
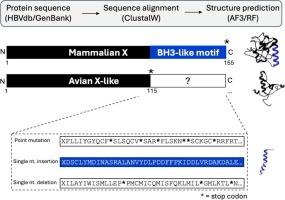
Chronic hepatitis B virus (HBV) infection is a leading cause of hepatocellular carcinoma (HCC). HBV produces an X protein (HBx) that is heavily linked to HCC, a similar finding seen across the Orthohepadnaviruses. There is an analogous, but truncated form of the X protein in Avihepadnaviruses, but no HCC is seen in infected avian species. This raises questions about the differences between mammalian and avian X proteins and their potential roles in carcinogenesis. Here we explore this question from a structural perspective of X proteins across mammalian and avian hepadnaviruses to determine whether there are any structural features linking these interesting observations. We first compile sequences to create consensus sequences with which to align with each other and subsequently to input into modeling programs, RoseTTAFold (RF) and AlphaFold3 (AF3). Comparative analyses show that mammalian X proteins are longer (154aa and 141aa, respectively for HBx and WHx) and have distinct domains in their C-terminal region, including a BH3-like domain that has been linked with many cancer pathways. Avian X proteins, however, are truncated with a similarly located stop codon. This truncation rids the protein of the BH3-like domain. We sought to find this domain by theoretical read-through or frameshifts and indeed, an analogous BH3-like domain is found that can similarly bind the BH3 target Bcl-2. Based on this work, it may be evolutionarily plausible that a single-nucleotide insertion led to the stop codon and BH3-domain loss that may account for the lack of cancers seen with Avihepadnaviruses.
跨物种X蛋白的比较结构见解和“丢失的”bh3样结构域可能解释鸟类肝细胞癌的缺失
慢性乙型肝炎病毒(HBV)感染是导致肝细胞癌(HCC)的主要原因。HBV产生一种与HCC密切相关的X蛋白(HBx),这在正肝病毒中也有类似的发现。在鸟肝病毒中也有类似的,但被截断的X蛋白形式,但在感染的禽类物种中未见HCC。这就提出了关于哺乳动物和鸟类X蛋白之间的差异及其在致癌作用中的潜在作用的问题。在这里,我们从哺乳动物和鸟类肝炎病毒X蛋白的结构角度探讨了这个问题,以确定是否有任何结构特征将这些有趣的观察联系起来。我们首先编译序列,以创建共识序列,与彼此对齐,随后输入到建模程序,RoseTTAFold (RF)和AlphaFold3 (AF3)。对比分析表明,哺乳动物的X蛋白更长(HBx和WHx分别为154aa和141aa),并且在其c端区域具有不同的结构域,包括与许多癌症途径相关的bh3样结构域。然而,禽类X蛋白被类似位置的终止密码子截断。这种截断使蛋白质脱离bh3样结构域。我们试图通过理论通读或帧移来找到这个结构域,并且确实发现了一个类似BH3的结构域,它可以类似地结合BH3的靶标Bcl-2。基于这项工作,单核苷酸插入导致停止密码子和bh3结构域的丢失可能是进化上合理的,这可能解释了阿维肝病毒没有癌症的原因。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Infection Genetics and Evolution
医学-传染病学
CiteScore
8.40
自引率
0.00%
发文量
215
审稿时长
82 days
期刊介绍:
(aka Journal of Molecular Epidemiology and Evolutionary Genetics of Infectious Diseases -- MEEGID)
Infectious diseases constitute one of the main challenges to medical science in the coming century. The impressive development of molecular megatechnologies and of bioinformatics have greatly increased our knowledge of the evolution, transmission and pathogenicity of infectious diseases. Research has shown that host susceptibility to many infectious diseases has a genetic basis. Furthermore, much is now known on the molecular epidemiology, evolution and virulence of pathogenic agents, as well as their resistance to drugs, vaccines, and antibiotics. Equally, research on the genetics of disease vectors has greatly improved our understanding of their systematics, has increased our capacity to identify target populations for control or intervention, and has provided detailed information on the mechanisms of insecticide resistance.
However, the genetics and evolutionary biology of hosts, pathogens and vectors have tended to develop as three separate fields of research. This artificial compartmentalisation is of concern due to our growing appreciation of the strong co-evolutionary interactions among hosts, pathogens and vectors.
Infection, Genetics and Evolution and its companion congress [MEEGID](http://www.meegidconference.com/) (for Molecular Epidemiology and Evolutionary Genetics of Infectious Diseases) are the main forum acting for the cross-fertilization between evolutionary science and biomedical research on infectious diseases.
Infection, Genetics and Evolution is the only journal that welcomes articles dealing with the genetics and evolutionary biology of hosts, pathogens and vectors, and coevolution processes among them in relation to infection and disease manifestation. All infectious models enter the scope of the journal, including pathogens of humans, animals and plants, either parasites, fungi, bacteria, viruses or prions. The journal welcomes articles dealing with genetics, population genetics, genomics, postgenomics, gene expression, evolutionary biology, population dynamics, mathematical modeling and bioinformatics. We also provide many author benefits, such as free PDFs, a liberal copyright policy, special discounts on Elsevier publications and much more. Please click here for more information on our author services .
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


