Shotgun Metagenomics Identifies in a Cross-Sectional Setting Improved Plaque Microbiome Biomarkers for Peri-Implant Diseases
Abstract
Aim
This observational study aimed to verify and improve the predictive value of plaque microbiome of patients with dental implant for peri-implant diseases.
Materials and Methods
Patients were included in one of the following study groups according to the health status of their dental implants: (a) healthy, (b) affected by mucositis and (c) affected by peri-implantitis. From each patient, submucosal plaque microbiome samples were collected from the considered dental implant and from a contralateral healthy implant/tooth. After shotgun metagenomic sequencing, the plaque microbiome was profiled taxonomically and functionally with MetaPhlAn 4 and HUMAnN 3, respectively. Taxonomic and functional profiles were fed into machine-learning models, which were then evaluated with cross-validation to assess the extent to which the plaque microbiome could be used to pinpoint peri-implant diseases.
Results
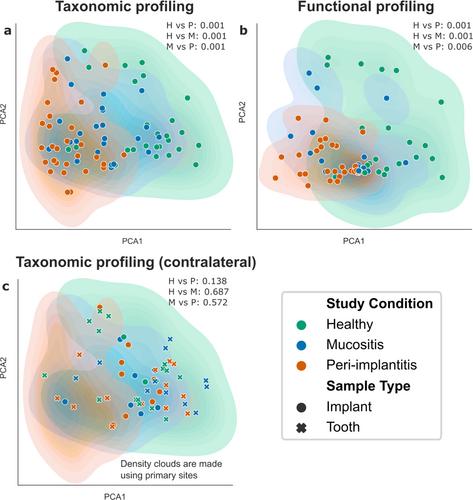
Shotgun metagenomics sequencing was performed for a total of 158 samples spanning 102 individuals. Four-hundred and forty-seven prokaryotic species were identified as part of the peri-implant microbiome, 34% of which were currently uncharacterized species. At the community level, the peri-implant microbiome differed according to the health status of the implant (p ≤ 0.006 for all pairwise comparisons) but this was site-specific, as healthy contralateral sites showed no discriminating microbiome features. Peri-implantitis microbiomes further showed lower inter-subject variability than healthy plaque microbiomes (p < 0.001), while mucositis-associated microbiomes were in the middle of the continuum between health and peri-implantitis. Each health condition was associated with a strong signature of taxonomic and functional microbiome biomarkers (log10 LDA score ≥ 2.5), 30% and 13% of which represented uncharacterized microbial functions and unknown species, respectively. Distinct Fusobacterium nucleatum clades were associated with implant status, highlighting the subspecies F. nucleatum′s functional and phenotypic diversity. Machine-learning models trained on taxonomic or functional plaque microbiome profiles were highly accurate in differentiating clinical groups (AUC = 0.78–0.96) and highlighted the extent to which the peri-implant microbiome is associated with peri-implant clinical parameters (AUC = 0.79–0.87).
Conclusions
Plaque microbiome profiling with shotgun metagenomics revealed consistent associations between microbiome composition and peri-implant diseases. In addition to pointing to peri-implant-associated microbes, warranting further mechanistic studies, we showed high-resolution plaque microbiome evaluation via metagenomics as an effective tool. Its utility within protocols for clinical management of peri-implant diseases should be explored in the future.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


