Biochemical analysis of PD-L1 ubiquitination by CRL3SPOP, ARIH1, and NEDD4 family ubiquitin ligases
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
As a key immune checkpoint ligand, PD-L1 is a critical target in cancer immunotherapy. While multiple E3 ubiquitin ligases including CRL3SPOP, ARIH1, and NEDD4 have been implicated in PD-L1 degradation, the precise enzymatic mechanisms remain unclear. In this study, we systematically compared the enzymatic activities of CRL3SPOP, ARIH1, and NEDD4 ligases toward the cytoplasmic domain of PD-L1 through in vitro reconstitution with purified components. ARIH1, rather than CRL3SPOP, independently ubiquitinates PD-L1. We reveal a mechanism where ARIH1 acts as a substrate receptor and cooperates with CRLs to catalyze PD-L1 ubiquitination. We also biochemically validated the E3 ligase activity of the NEDD4 family E3s toward PD-L1. By using liposomes in the enzymatic assays, we show that phosphorylation enhances PD-L1 ubiquitination through disrupting its membrane association. Our study provides further biochemical insights into PD-L1 ubiquitination, which advances our understanding of the molecular details of PD-L1 regulation.
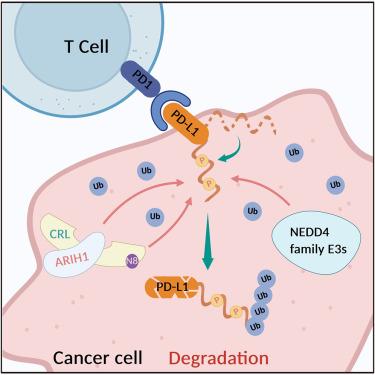
CRL3SPOP、ARIH1和NEDD4家族泛素连接酶对PD-L1泛素化的生化分析
作为一种关键的免疫检查点配体,PD-L1是肿瘤免疫治疗的重要靶点。虽然包括CRL3SPOP、ARIH1和NEDD4在内的多种E3泛素连接酶与PD-L1降解有关,但确切的酶促机制尚不清楚。在本研究中,我们系统地比较了CRL3SPOP、ARIH1和NEDD4连接酶对PD-L1细胞质结构域的酶活性。ARIH1,而不是CRL3SPOP,独立泛素化PD-L1。我们揭示了ARIH1作为底物受体并与CRLs合作催化PD-L1泛素化的机制。我们还生化验证了NEDD4家族E3对PD-L1的连接酶活性。通过使用脂质体进行酶分析,我们发现磷酸化通过破坏其膜结合来增强PD-L1泛素化。我们的研究为PD-L1泛素化提供了进一步的生化见解,这促进了我们对PD-L1调控的分子细节的理解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


