First report of a carbapenem-resistant Serratia sarumanii clinical strain co-harboring blaKPC-2 and blaNDM-1 genes in China
IF 5.8
Q1 MICROBIOLOGY
引用次数: 0
Abstract
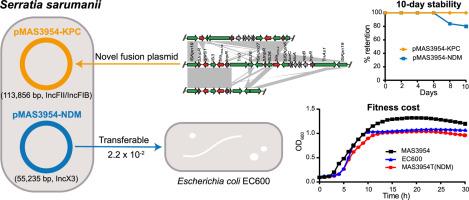
Carbapenem-resistant Enterobacteriaceae (CRE), particularly those co-harboring multiple carbapenemase genes, pose a significant global health threat. However, the coexistence of blaKPC-2 and blaNDM-1 in Serratia sarumanii has not been previously reported. This study aims to report and characterize the first carbapenem-resistant S. sarumanii (CRSS) clinical strain MAS3954 in China co-harboring blaKPC-2 and blaNDM-1, focusing on its genetic characteristics, plasmid stability, and transfer potential. Whole-genome analysis revealed that the blaKPC-2 and blaNDM-1 were located on two distinct plasmids. Plasmid pMAS3954-KPC (113,856 bp, IncFII/IncFIB) exhibited low similarity (<66%) to known plasmids, indicating a novel fusion event between pKPC-h2 and S. marcescens chromosome, while pMAS3954-NDM (55,235 bp, IncX3) was highly conserved (100% identity/coverage). Conjugation experiments showed that the blaNDM-1 was transferable, while blaKPC-2 was not. During 10 days of continuous passage, the genetic context of blaNDM-1 was gradually excised from the plasmids after the 8th day, whereas they maintained 100% retention for blaKPC-2. S. sarumanii MAS3954 was multidrug-resistant (MDR), including carbapenems, β-lactams, β-lactam/β-lactamase inhibitors, trimethoprim-sulfamethoxazole, tetracycline, nitrofurantoin, colistin, and fluoroquinolones, but remained susceptible to certain aminoglycosides and tigecycline. Phylogenomic analysis identified a distinct clade for S. sarumanii MAS3954, diverging notably from other strains. Comparison of resistance genes further highlighted the unique co-harboring of blaKPC-2 and blaNDM-1 in MAS3954, absent in other strains. To our knowledge, this study represents the first characterization of clinical S. sarumanii strain co-harboring blaKPC-2 and blaNDM-1. The findings highlight the need for enhanced surveillance and infection control to prevent the spread of these MDR strains in healthcare settings.
中国首次报道一株同时携带blaKPC-2和blaNDM-1基因的耐碳青霉烯萨鲁曼沙雷菌临床菌株
碳青霉烯耐药肠杆菌科(CRE),特别是那些共同携带多个碳青霉烯酶基因的肠杆菌,对全球健康构成重大威胁。然而,blaKPC-2和blaNDM-1在萨鲁曼沙雷氏菌中的共存尚未见报道。本研究旨在报道和鉴定中国首个含有blaKPC-2和blaNDM-1基因的耐碳青霉烯sarumanii (CRSS)临床菌株MAS3954,重点研究其遗传特性、质粒稳定性和转移潜力。全基因组分析显示blaKPC-2和blaNDM-1位于两个不同的质粒上。质粒pMAS3954-KPC (113,856 bp, IncFII/IncFIB)与已知质粒的相似性较低(<66%),表明pKPC-h2与S. marcescens染色体之间存在新的融合事件,而pMAS3954-NDM (55,235 bp, IncX3)高度保守(100%同源/覆盖率)。偶联实验表明blaNDM-1具有可转移性,而blaKPC-2不具有可转移性。在10天的连续传代中,blaNDM-1的遗传背景在第8天后逐渐从质粒中去除,而blaKPC-2的遗传背景则保持100%的保留。S. sarumanii MAS3954对碳青霉烯类、β-内酰胺类、β-内酰胺/β-内酰胺酶抑制剂、甲氧苄啶-磺胺甲恶唑、四环素、呋喃托因、粘菌素和氟喹诺酮类多药耐药,但对某些氨基糖苷类和替加环素敏感。系统基因组学分析鉴定出萨鲁曼链球菌MAS3954有一个独特的分支,与其他菌株明显不同。抗性基因的比较进一步突出了MAS3954中blaKPC-2和blaNDM-1的共藏性,这在其他菌株中不存在。据我们所知,这项研究首次表征了临床萨鲁曼链球菌共携带blaKPC-2和blaNDM-1的菌株。这些发现强调需要加强监测和感染控制,以防止这些耐多药菌株在卫生保健环境中传播。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Current Research in Microbial Sciences
Immunology and Microbiology-Immunology and Microbiology (miscellaneous)
CiteScore
7.90
自引率
0.00%
发文量
81
审稿时长
66 days
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


