Distinct tau filament folds in human MAPT mutants P301L and P301T
IF 10.1
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
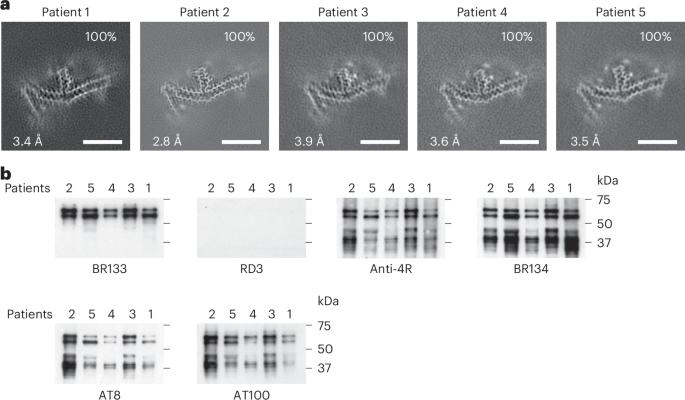
Mutations in MAPT, the tau gene, give rise to frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17), with abundant filamentous tau inclusions in brain cells. Mutations that encode missense variants of residue P301 are the most common and result in the formation of filamentous inclusions made of mutant four-repeat tau. Here we report the cryo-electron microscopy structures of tau filaments from five individuals belonging to three different families with mutation P301L and from one individual from a family with mutation P301T. A distinct three-lobed tau fold resembling the two-layered fold of Pick’s disease was present in the individuals with P301L tau. Two different tau folds were found in the individual with mutation P301T, the less abundant of which was a variant of the three-lobed fold. The major P301T tau fold was V-shaped, with partial similarity to the four-layered tau folds of corticobasal degeneration and argyrophilic grain disease. Schweighauser, Shi, Murzin and colleagues report cryo-EM structures of tau filaments from individuals with P301L or P301T MAPT mutations. P301L tau filaments adopted a distinct three-lobed fold, while P301T filaments had either a variant of the three-lobed fold or a V-shaped fold.

人类MAPT突变体P301L和P301T中不同的tau丝折叠
tau基因MAPT的突变可引起与17号染色体(FTDP-17)相关的额颞叶痴呆和帕金森病,在脑细胞中存在大量丝状tau内含物。编码残基P301错义变体的突变是最常见的,并导致由突变的四重复tau构成的丝状内含物的形成。在这里,我们报告了来自三个不同突变P301L家族的五个个体和来自突变P301T家族的一个个体的tau蛋白细丝的低温电镜结构。具有P301L tau的个体中存在明显的三叶tau褶皱,类似于匹克病的双层褶皱。在突变P301T的个体中发现了两个不同的tau折叠,其中较少的是三叶折叠的变体。P301T的主要tau褶皱呈v形,部分类似于皮质基底变性和嗜银性谷物病的四层tau褶皱。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


