Proteomic profiling of serum small extracellular vesicles predicts post-COVID syndrome development
IF 3.8
3区 医学
Q2 IMMUNOLOGY
引用次数: 0
Abstract
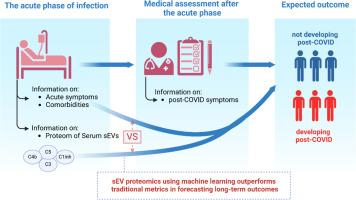
Post-COVID syndrome affects 10–35 % of COVID-19 patients, and up to 85 % of hospitalized individuals, underscoring the need for early identification of high-risk cases. We hypothesized that the proteomic profile of serum small extracellular vesicles (sEVs) obtained during acute SARS-CoV-2 infection could predict post-COVID syndrome.
Serum samples from 59 patients, stratified as asymptomatic, moderate, or severe, were analyzed. sEVs were isolated, characterized by electron microscopy, nanoparticle tracking, and flow cytometry, then profiled via LC-MS.
Classification models integrating comorbidities, acute symptoms, and sEV proteomics distinguished the three groups, with sEV data outperforming conventional measures. Of 620 identified proteins, 30 showed significant differences between symptomatic and asymptomatic patients, including 12 linked to complement activation. ELISA confirmed LC-MS results that serum sEVs of post-COVID patients had altered C1 inhibitor, C3, and C5 levels.
These results suggest that sEV-based proteomics can enable earlier detection and more targeted follow-up for individuals at risk of post-COVID syndrome.
血清细胞外小泡的蛋白质组学分析预测covid后综合征的发展。
COVID-19后综合征影响10- 35% %的COVID-19患者和高达85% %的住院患者,这突出了早期识别高风险病例的必要性。我们假设急性SARS-CoV-2感染期间获得的血清小细胞外囊泡(sev)的蛋白质组学特征可以预测covid后综合征。对59例无症状、中度或重度患者的血清样本进行分析。分离sev,通过电子显微镜、纳米颗粒跟踪和流式细胞术进行表征,然后通过LC-MS进行分析。综合合并症、急性症状和sEV蛋白质组学的分类模型区分了三组,sEV数据优于常规测量。在620个确定的蛋白质中,30个在有症状和无症状的患者之间显示出显著差异,其中12个与补体激活有关。ELISA证实LC-MS结果,新冠肺炎后患者血清sev水平发生改变,C1抑制剂、C3和C5水平发生改变。这些结果表明,基于sev的蛋白质组学可以使有后冠状病毒综合征风险的个体能够更早地发现和更有针对性的随访。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Clinical immunology
医学-免疫学
CiteScore
12.30
自引率
1.20%
发文量
212
审稿时长
34 days
期刊介绍:
Clinical Immunology publishes original research delving into the molecular and cellular foundations of immunological diseases. Additionally, the journal includes reviews covering timely subjects in basic immunology, along with case reports and letters to the editor.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


